Computational Oncology
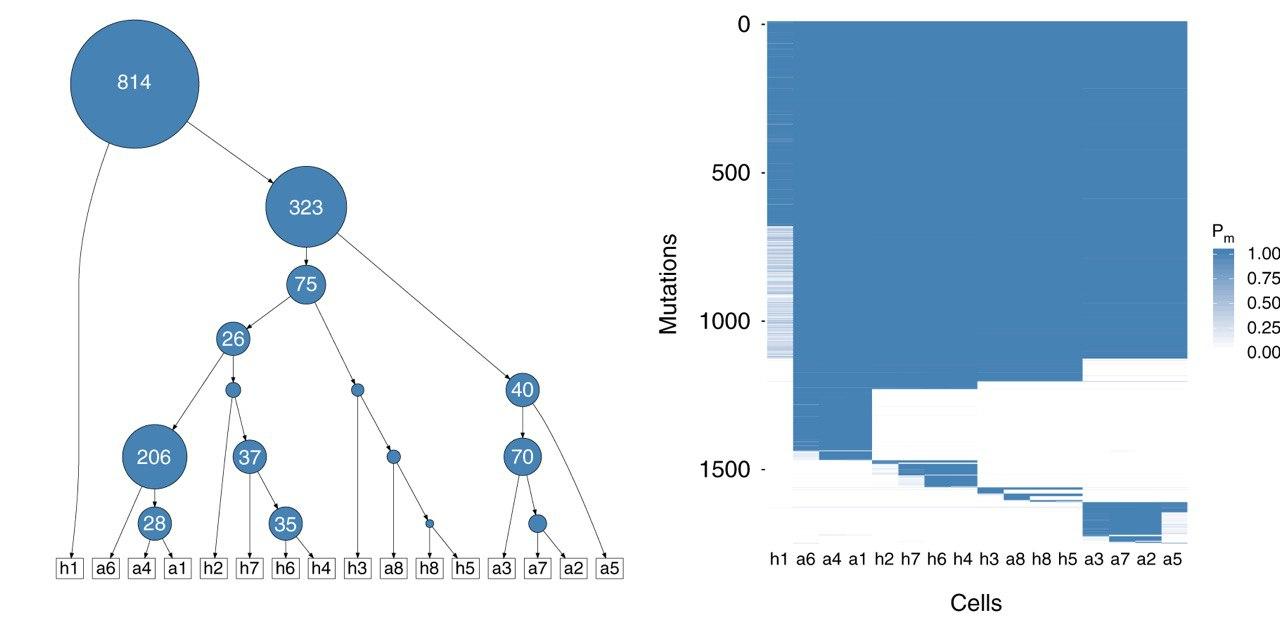
Our cancer-related research includes the analysis, integration, and interpretation of molecular profiles of tumors. Our goal is to support the diagnosis and treatment of cancer. For example, we have developed methods for improved mutation calling from bulk and single-cell tumor sequencing data that allow for detecting genetic variants that occur at low frequencies within a tumor.
For the interpretation of mutations found in tumors, we develop methods that discriminate driver from passenger mutations, i.e., mutations that confer a selective advantage to the cancer cell and promote its clonal expansion from selectively neutral and therefore clinically irrelevant mutations.
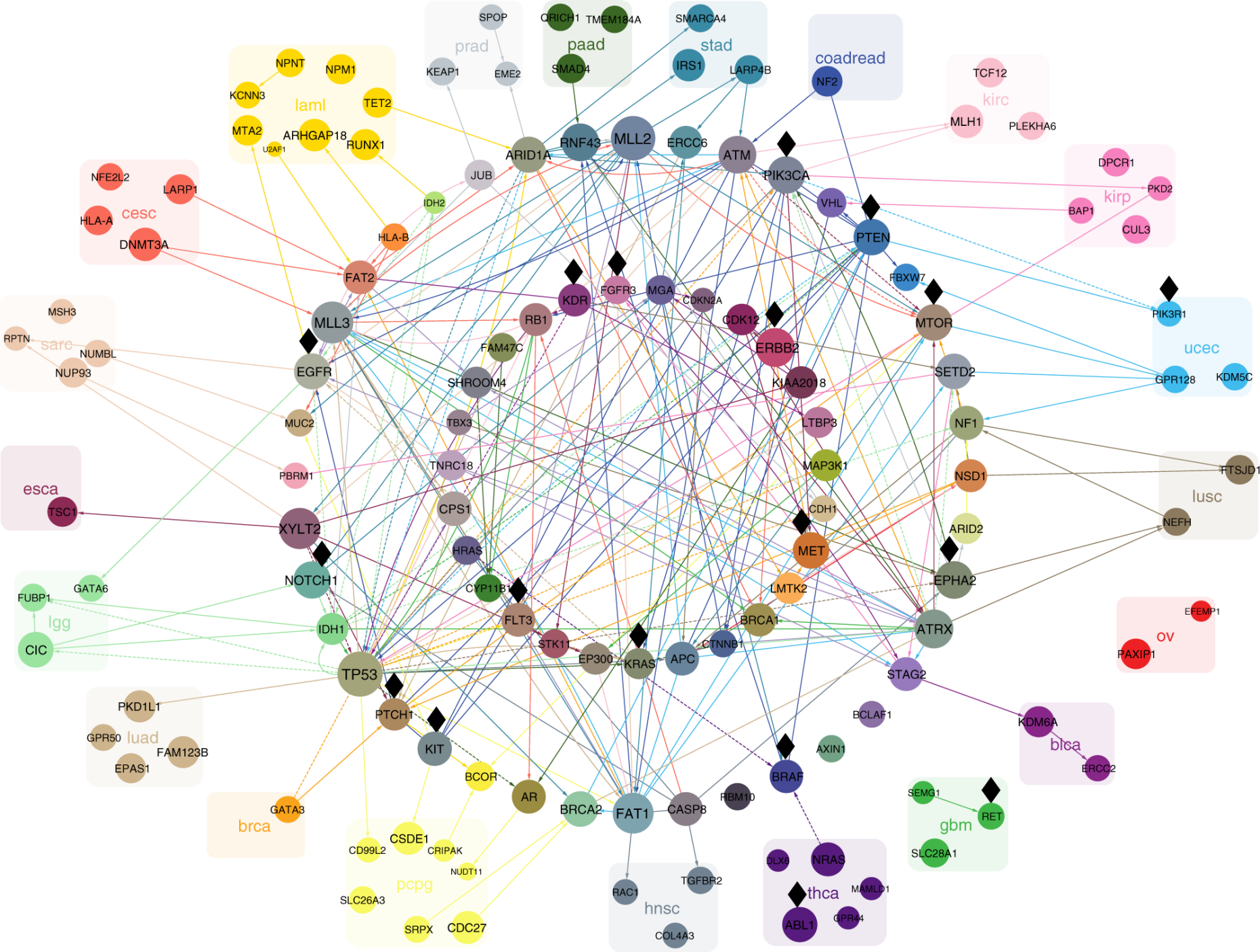
In order to identify groups of genes, or pathways, that are involved in cancer, we build statistical models to identify sets of mutually exclusive cancer mutations, which are likely to interact and to serve a biological function that is critical for tumor survival and growth. Similarly, pairs of synthetic lethal mutations can indicate novel therapeutic targets.
Selected references:
- external page Tree inference for single-cell data
Jahn et al., Genome Biol, 2016
- external page Mutational interactions define novel cancer subgroups
Kuipers et al., Genome Res, 2018 - external page Single-cell mutation identification via phylogenetic inference
Singer et al., Nat Commun, 2018