Networks and Perturbations
Interactions among molecules, cells, and individuals determine the properties and the behavior of a biological system. These interactions give rise to various biological networks and their mathematical representations as graphs. We use biological networks for integrating different data types. For example, cancer cells can be profiled for their DNA, RNA, and protein composition, and we use gene regulatory networks and protein-protein interaction networks to combine the evidence from all molecular measurements.
Molecular changes, such as mutations, can be regarded as perturbations of a biological network. The connectivity of the network is used to predict the effect of the perturbation on the system. We employ graph diffusion techniques to estimate the system-level effect of individual patterns of perturbations, such as mutational profiles or differentially expressed genes of tumor cells.
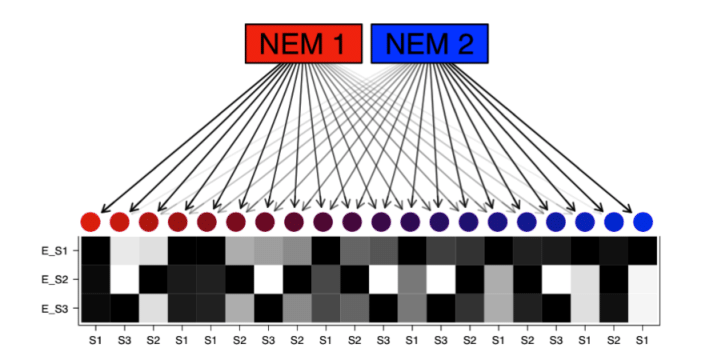
Network perturbations also reveal information on the structure of the underlying network, and we build computational methods for learning networks from perturbation data. For example, we have developed several nested effects models for reconstructing intracellular signaling networks from RNA interference and CRISPR-Cas9 screening data.
Selected references:
- external page Single cell network analysis with a mixture of Nested Effects Models
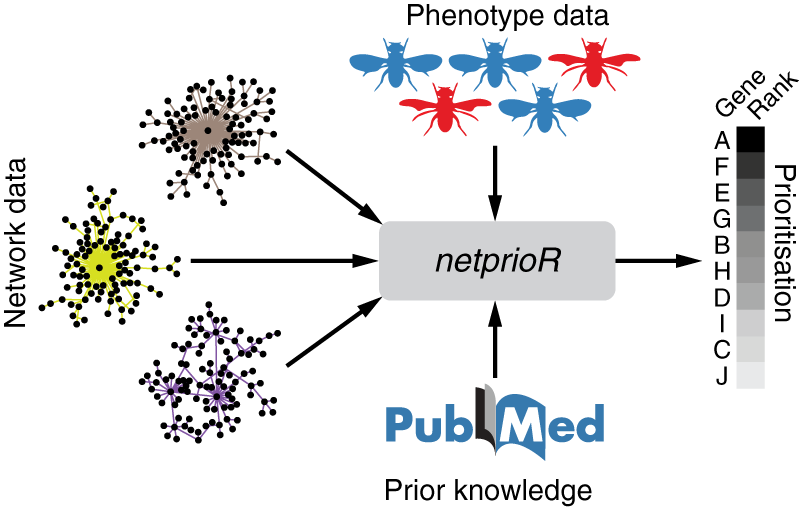
Pirkl et al., Bioinformatics, 2018 - external page netprioR: a probabilistic model for integrative hit prioritisation of genetic screens
Schmich et al., Stat Appl Genet Mol, 2019 - external page Inferring signalling dynamics by integrating interventional with observational data
Cardner et al., Bioinformatics, 2019