Epigenetic pathways in cancer
There is increasing evidence that mis-regulation of epigenetic mechanisms can leads to cancer development. Human tumors often show mutations in genes encoding chromatin factors and particularly in components of the cellular memory machinery. In flies, downregulation of Polyhomeotic (Ph), a member of the PRC1 PcG protein complex, leads to neoplastic growth of imaginal discs, as this epigenetic regulator acts as a tumor suppressor. We utilized an inducible tumor system to test whether the development of cancer is associated with the gradual acquisition of genetic alterations leading to a progressive increase in malignancy. To this purpose, we did a time-course analysis of whole-genome deep sequencing of the tumor tissue and compared them to the genetically matched control tissues. Our results demonstrate that genome stability is not inevitably affected during sustained tumor growth in Drosophila. Hence, derailment of cellular and epigenetic networks governing signaling pathways and growth control is sufficient to drive tumorigenesis in Drosophila.
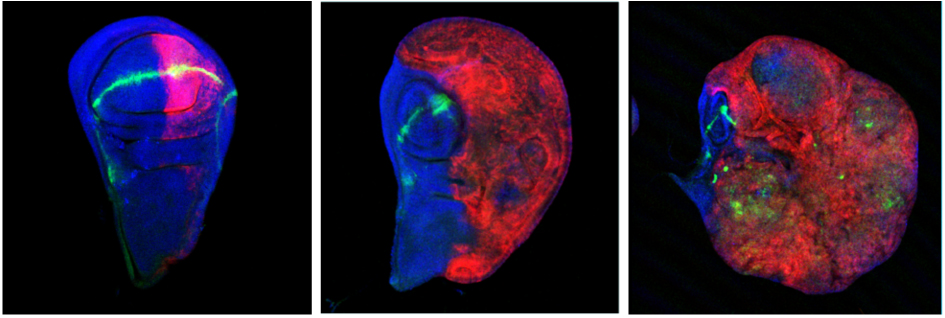
Inducing reversible epigenetic tumors
Polyhomeotic tumor cells transform from a larval to an embryonic signature, driven by the expression of a newly identified oncogene. When abolishing the expression of this gene, the tumor tissue regress. Interestingly, the oncogene appears to activate signaling pathways which prevent the tumor cells to differentiate. When artificially forcing the cells to differentiate, however, by expression of a transcription factor involved in differentiation, they lose their tumorigenic activity.
An intrinsic tumor eviction mechanism in Drosophila
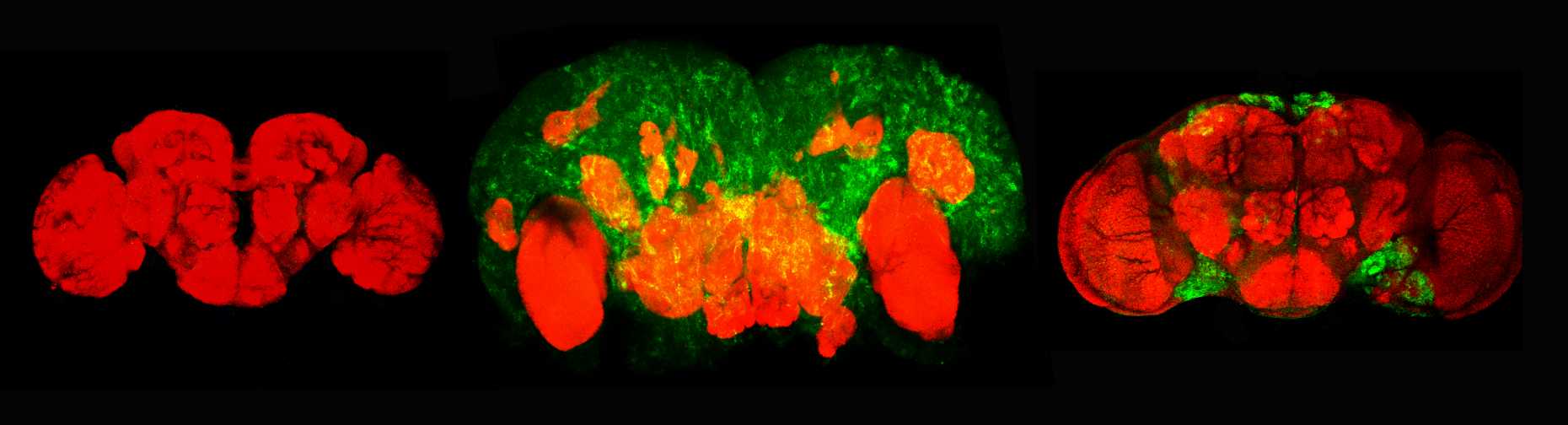
Flies evolved an innate tumor suppression mechanism. At metamorphosis the steroid hormone ecdysone alters neoplastic growth of PcG tumors into a non-tumorigenic condition. Subsequently, these cells become eliminated during the adult stage. The hormone Ecdysone exerts this function by inducing a heterochronic network encompassing the activation of the microRNA lethal-7 which suppresses the tumor driver chinmo. Interestingly, this pathway can also promote remission of brain tumors formed in brain tumor mutants, revealing a restraining of neoplastic growth of different tumor types during metamorphosis. Given the conserved role of let-7, the identification and molecular characterization of this tumor eviction mechanism in flies might provide important clues toward the exploitation of related pathways for human tumor therapy.
Elimination of brain tumor cells by the let-7 pathway. Left and middle: normal adult fly brain and fly brain with brat tumor cells (green). Right: after activation of let-7 cascade tumor cells become eliminated in the adult brain.
Since several examples of human tumors with epigenetic alterations have been identified, it will be interesting to test whether our work with a simpler model organism will reveal in the future comparable pathways in the more complex human disease setting. Indeed, driving cancer cells from a highly proliferative, non-differentiated state into a dead-end, differentiated state is a little pursued strategy termed “differentiation therapy”. Work with flies might unravel basic concepts and mechanisms to achieve such cellular transformations more directly and efficiently. It may take time for a therapeutic targeting, but a basic understanding of the process is the necessary seed for such advances.