Synthetic Immunology
We are applying a number of genome engineering tools (e.g., CRISPR-Cas9) to reprogram immune cells for applications related to biotechnology and cellular immunotherapy.
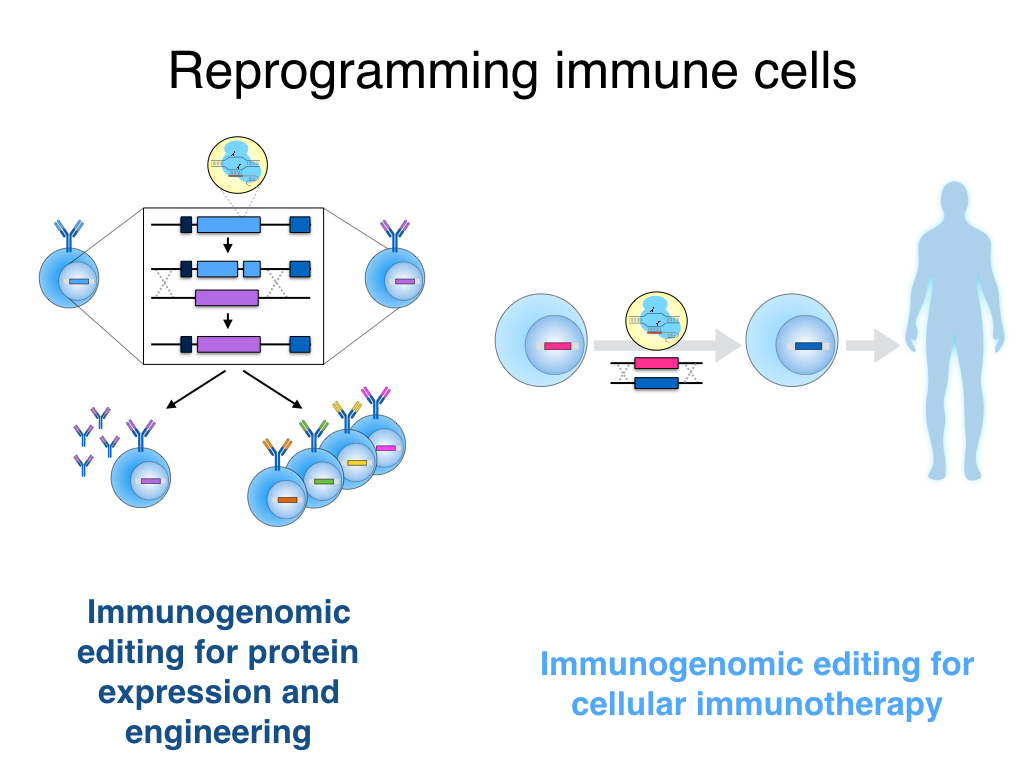
Reprogramming immune cells for protein expression and engineering
In one of our research areas, we focus on the development of technologies for protein expression and engineering using advanced methods in immunogenomic reprogramming.
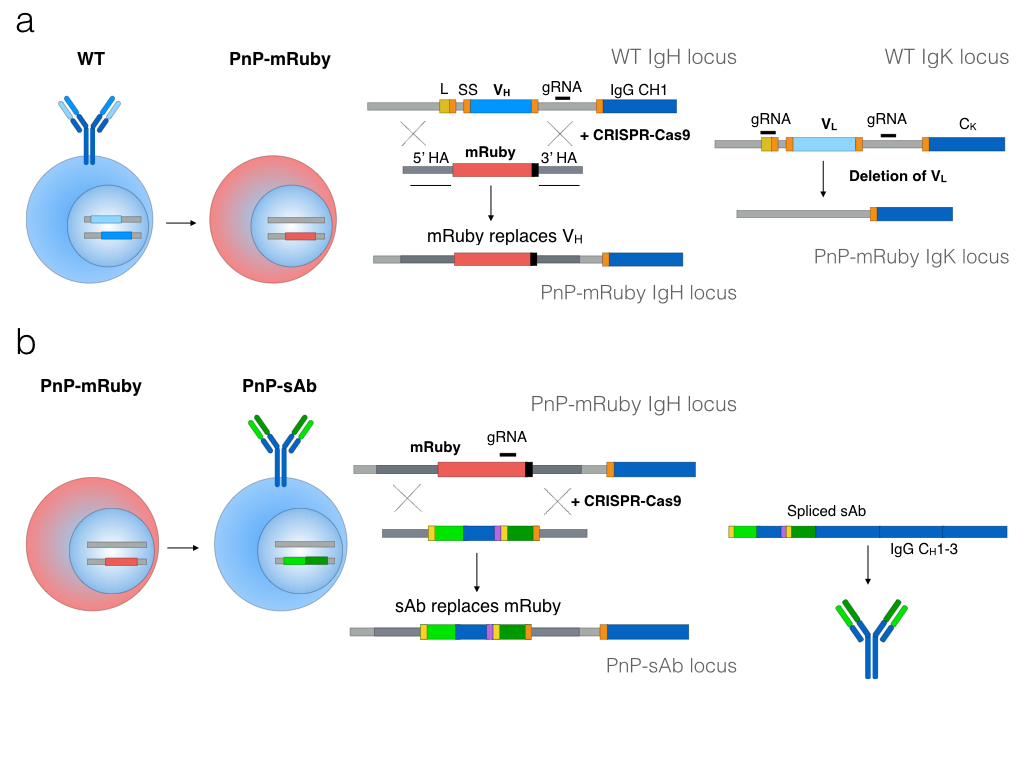
Specifically, we have recently developed Plug-and-(dis)play hybridoma cells, which is a platform for rapid reprogramming of antibody specificity by immunogenomic engineering (Pogson*, Parola* et al., Nature Communications, 2016).
Hybrdomas are stable, rapidly-proliferating cell lines widely utilized for antibody screening and production, which are generated by fusing primary mouse B cells with myelomas tumor cells. Antibody specificity of a hybridoma clone is determined by the immunoglobulin sequence of the primary B cell. We use CRISPR-Cas9 to generate double-stranded breaks in immunoglobulin loci, enabling deletion of the native variable light chain and replacement of the endogenous variable heavy chain with a fluorescent reporter protein (mRuby) (Fig. 1). New antibody genes are introduced by Cas9-targeting of mRuby for replacement with a donor construct encoding a light chain and a variable heavy chain, resulting in full-length antibody expression. Since hybridomas surface express and secrete antibodies, reprogrammed cells are isolated using flow cytometry and cell culture supernatant is used for antibody production. Plug-and-(dis)play hybridomas can be reprogrammed with only a single transfection and screening step.
Reprogramming immune cells for cellular immunotherapy
The rapid advances in genome editing technologies are stimulating tremendous excitement for developing advanced cellular immunotherapies. To date, proof-of-concept examples of genome editing in immune cells have shown targeted insertion, deletion, or point mutation of genes. However, what has yet to be reported is the exchange of entire genes, particularly those of polymorphic immunogenomic loci. The ability to exchange immunogenomic alleles would enable the reprogramming of immune cell specificity.
One important immunogenomic region is the MHC locus; these genes are critically important in allogeneic hematopoietic stem cell (HSC) transplantation procedures, where MHC matching between the donor and the host is a primary determinant of transplant success. While the editing of human HSCs has traditionally been difficult, new advances are greatly improving efficacy to the levels required for clinical application. In fact, very recent developments indicate genome editing of human HSCs will shortly gain clinical approval in Europe for Severe Combined Immune Deficiency disease. However, what remains a major challenge is the need to still obtain proper MHC matching of donor and host.
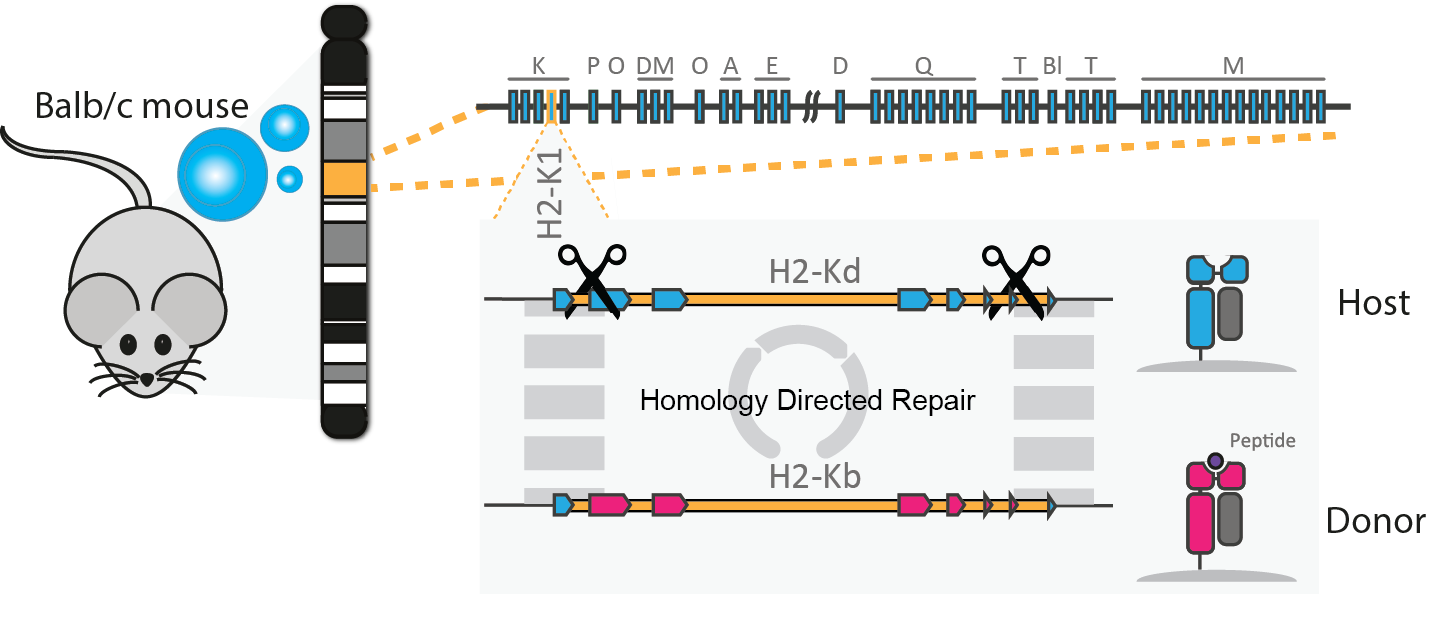
As a proof of concept for MHC reprogramming, we targeted a murine MHC locus (H2-K1) with our approach of CRISPR-Cas9 assisted cassette exchange (Kelton et al., Scientific Reports, 2017).
In murine-derived antigen-presenting cell lines, we introduced double strand breaks with a multiplex-targeting Cas9 approach. Providing repair templates, we enabled homology directed repair (HDR) of the double-stranded break. Thus, we were able to effectively replace a MHC-I allele (H2-Kd) of ~3.4 kb with a highly similar orthogonal allele (H2-Kb). We validated the altered cellular phenotype by the surface expression of the new MHC alleles, using fluorescence associated cell sorting(and a T cell activation assay, highlighting functional immune activity of the MHC exchanged cells.