Computational Methods
We develop computational methods for the modeling and simulation of signalling networks, cell-based tissue simulations, image processing and image-based modelling.
Cell-based Tissue Simulation Framework
We have developed LBIBCell, PolyHoop and SimuCell3D (all C++-based) to simulate tissue development in 2D and 3D at cellular resolution.
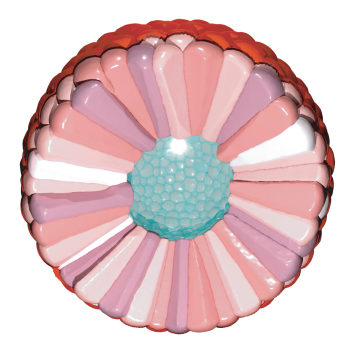
SimuCell3D: 3D Simulation of Tissue Mechanics with Cell Polarization
Steve Runser, Roman Vetter, and Dagmar Iber
external page Nature Computational Science (2024), external page News & Views, external page Tweetorial
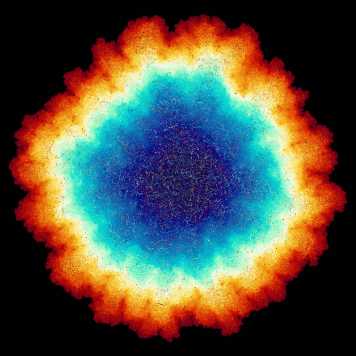
PolyHoop: Soft particle and tissue dynamics with topological transitions.
Roman Vetter, Steve Runser, and Dagmar Iber
external page Computer Physics Communications (2024), external page Tweetorial
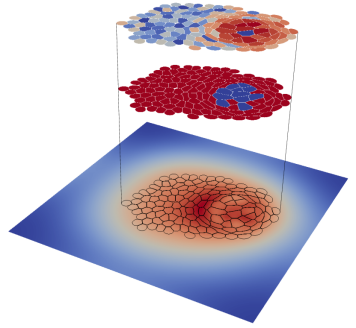
LBIBCell: A Cell-Based Simulation Environment for Morphogenetic Problems.
Tanaka S, Sichau D, Iber D, external page Bioinformatics, 2015.
external page Documentation; external page Download
Model Conversion and Parameter Estimation
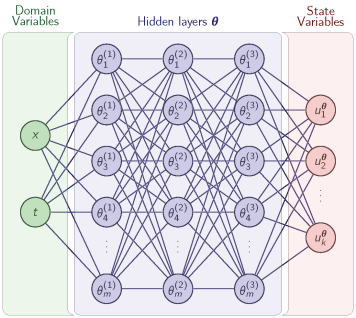
PINNverse: Accurate parameter estimation in differential equations from noisy data with constrained physics-informed neural networks.
Almanstötter M, Vetter R, Iber D, external page arXiv, 2025, external page Tweetorial
MOCCASIN: converting MATLAB ODE models to SBML.
Harold F. Gómez, Michael Hucka, Sarah M. Keating, German Nudelman, Dagmar Iber, Stuart C. Sealfon, external page Bioinformatics, 2016. external page Download
Image Processing
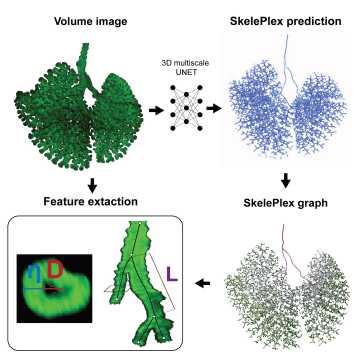
SkelePlex, a neural network-based image processing pipeline for quantification of bronchial trees.
Malte Mederacke, Kevin Yamauchi, et al., external page bioRxiv, 2025, external page Tweetorial
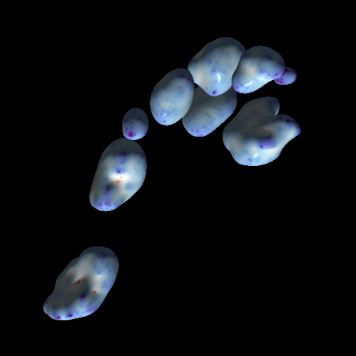
FollicleFinder: automated three-dimensional segmentation of human ovarian follicles.
Kevin Yamauchi, Marieke Biniasch, Leopold Franz, Harold Gómez, Christian De Geyter, Dagmar Iber, external page bioRxiv, 2022, external page Tweetorial
COMSOL
We are developing COMSOL applications for image-based modelling.
Simulating Organogenesis in COMSOL: Tissue Patterning with Directed Cell Migration
M Mederacke, C Yu, R Vetter and D Iber,
Proceedings of COMSOL Conference 2025 Amsterdam
Simulating Organogenesis in COMSOL: Tissue Mechanics
Peters, M and Iber, D,
external page Proceedings of COMSOL Conference 2017 Rotterdam
Simulating organogenesis in COMSOL: comparison of methods for simulating branching morphogenesis
LD Wittwer, M Peters, S Aland, D Iber
external page Proceedings of COMSOL Conference 2017 Rotterdam
Simulating organogenesis in COMSOL: Phase-field based simulations of embryonic lung branching morphogenesis
LD Wittwer, R Croce, S Aland, D Iber
external page Proceedings of COMSOL Conference 2016 Munich
Simulating Organogenesis in COMSOL: Image-based Modeling
Z Karimaddini, E Unal, D Menshykau, D Iber
external page Proceedings of COMSOL Conference, 2014
Simulating organogenesis in COMSOL: cell-based signaling models
J Vollmer, D Menshykau, D Iber
external page Proceedings of COMSOL Conference, 2013
Simulating Organogenesis in COMSOL: Parameter Optimization for PDE-based models
D Menshykau, S Adivarahan, P Germann, L Lermuzeaux, D Iber
external page Proceedings of COMSOL Conference, 2013
Simulating organogenesis with COMSOL: Deforming and interacting domains
D Menshykau, D Iber
external page Proceedings of COMSOL Conference 2012
Simulating organogenesis in COMSOL
P Germann, D Menshykau, S Tanaka, D Iber
external page Proceedings of COMSOL Conference, 2011