AAV-Perturb-seq
High-throughput in vivo CRISPR screening with high-resolution single-cell transcriptional phenotyping
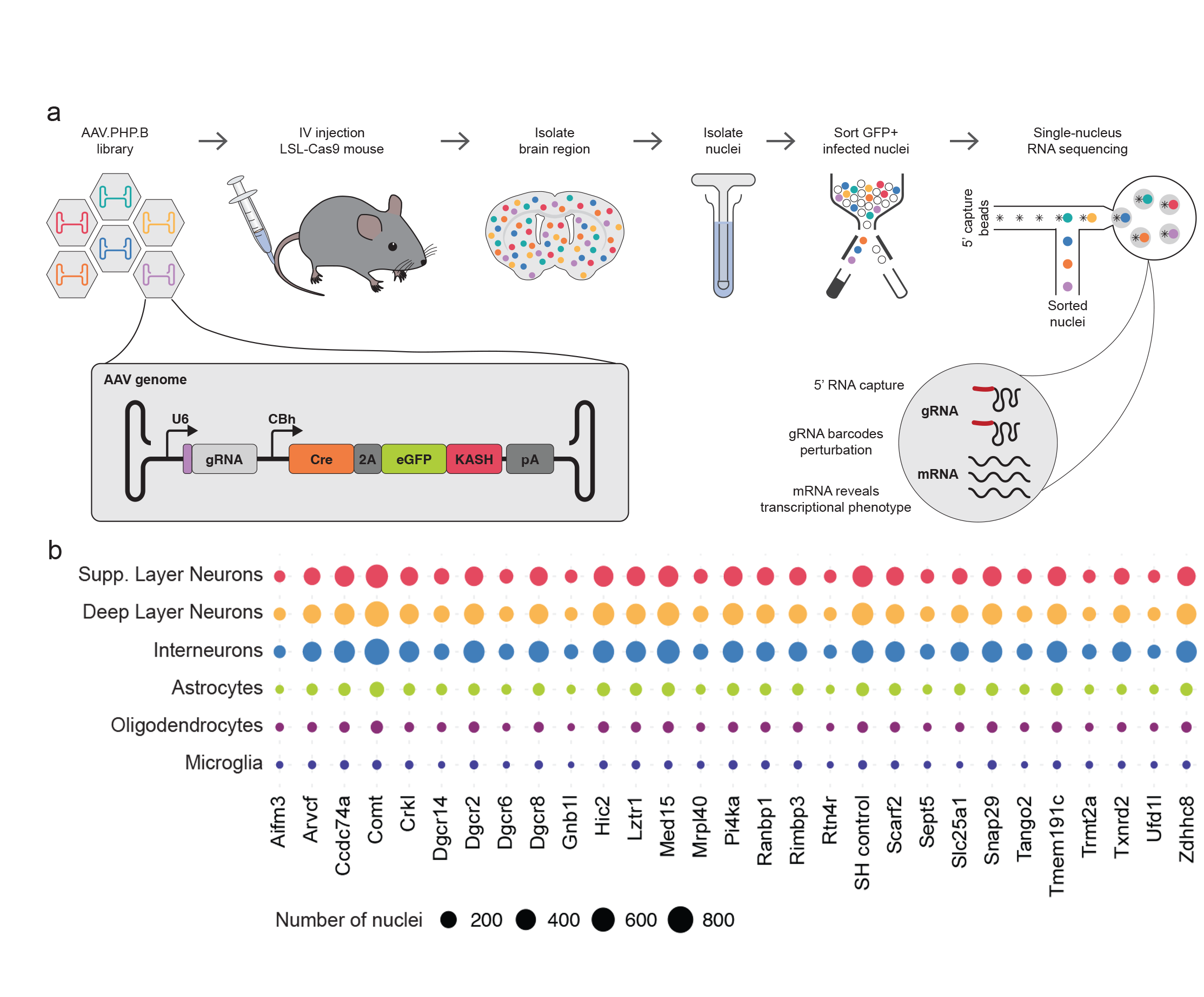
Understanding the function of genetic elements and how their disruption leads to disease is a central goal of biomedical research. Over the past few decades, the scientific community has tentatively associated thousands of genetic elements with disease, but identifying which ones are causal, understanding their function in health and disease, and developing targeted therapies has been much more challenging. This is in part due to the lack of high-throughput methodologies that link genotype to phenotype in model systems that recapitulate the physiology of cells and tissues in the body. To accelerate progress in this area, we developed AAV-Perturb-seq, a method that combines the flexibility of adeno-associated virus (AAV)-mediated in vivo delivery in animal models, the scalability of pooled CRISPR perturbation screening, and the resolution of single-cell transcriptomics. AAV-Perturb-seq involves producing a CRISPR guide RNA library targeting many genes of interest, packaging the library in one of the many available AAVs having tunable properties enabling them to infect certain cells and tissues of interest. Injecting the AAV CRISPR library into animal models then enables gene editing of the target genes in the cell and tissue types of interest. The modified cells or nuclei are then retrieved and analyzed by single-cell or single-nucleus RNA sequencing, enabling the high-throughput connection of genotype to transcriptional phenotype. AAV-Perturb-seq therefore enables detailed investigation of the effects of dozens of genetic perturbations across a range of cell types in
We used AAV-Perturb-seq to investigate the role of the genes involved in 22q11.2 deletion syndrome, which is caused by the loss of the chromosomal region 22q11.2. 22q11.2 deletion syndrome is the most common chromosomal deletion in humans and leads to heterogeneous clinical presentations including developmental defects, schizophrenia, and autism among many others. We used AAV-Perturb-seq to examine the function of 29 genes in the deleted region in the adult mouse brain and found that perturbation of only four genes resulted in transcriptional changes. Further analysis showed that three of these genes (Gnb1l, Dgcr8, and Dgcr14) influenced various biological pathways involved in neuron function. Our findings revealed potential contributors to neuronal dysfunction in 22q11.2 deletion syndrome and suggest that it might be possible to reverse some of the changes in gene expression directly in mature neurons.
AAV-Perturb-seq is a flexible method that can be applied broadly to investigate the role of genetic elements across a wide range of developmental time points, tissues, and cell types in high-throughput. In the future, AAV-Perturb-seq will accelerate progress toward identifying genetic elements underlying disease, understanding the function of these elements in living organisms, and discovering new therapeutic strategies.
Downloads and links
AAV-Perturb-seq, Experimental protocols
external page Santinha et al. Nature 2023
AAV-Perturb-seq code
external page Platt Lab GitHub
Deep sequencing data
external page Santinha et al. Nature 2023
external page GEO database
Plasmids (Addgene)
external page https://www.addgene.org/browse/article/28243551/
Further reading
Santinha et al. Nature 2023