Transcriptional programming with SiT-Cas12a

The picture shows a gene regulatory network. Each dot represents a gene and each line the functional interaction between distinct genes. Alteration of these connections are either cause or consequences of human diseases. The ability to recreate these functional interactions will allow scientist to better understand mechanisms of human diseases and to design more sophisticated therapeutic treatments. Now, our recent CRISPR platform provides a solution to manipulate and control these gene regulatory networks easily.
Multiplexed gene editing and transcriptional regulation by Cas12a and CRISPR arrays
Encoding of Cas12a and crRNA on a single mRNA
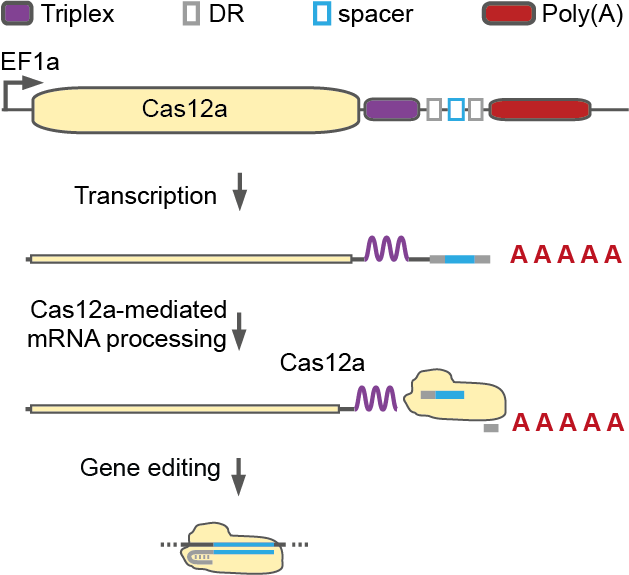
Schematic of a single transcript architecture containing both a Cas12a and a CRISPR array (Sit-Cas12a). The transcript encodes for: Cas12a (yellow rectangle); Triplex, a tertiary RNA structural motif (small purple rectangle); direct repeat (DR, grey square); spacer (blue square); and a polyadenylation sequence (Poly(A), red rectangle). After transcription and Cas12a mediated crRNA processing, the Triplex sequence stabilizes the transcript enabling concomitant Cas12a expression and gene editing.
Downloads and links
Deep sequencing data
external page Available on NCBI Sequence Read Archive (SRA)
Plasmids
external page Available from Addgene
The plasmids below have been deposited and are being processed by Addgene. The link above will take you to the Platt Lab Addgene page and the plasmids from the paper will be visable once available.
pCE048-SiT-Cas12a
pCE046-SiT-Cas12a-[Repr]
pCE059-SiT-Cas12a-[Activ]
pCI152-pLenti-SiT-Cas12a
pCI118-SiT-Cas12a-[Ind]
pCI108-SiT-Cas12a-[Cond]