Computer Control of Cell Populations
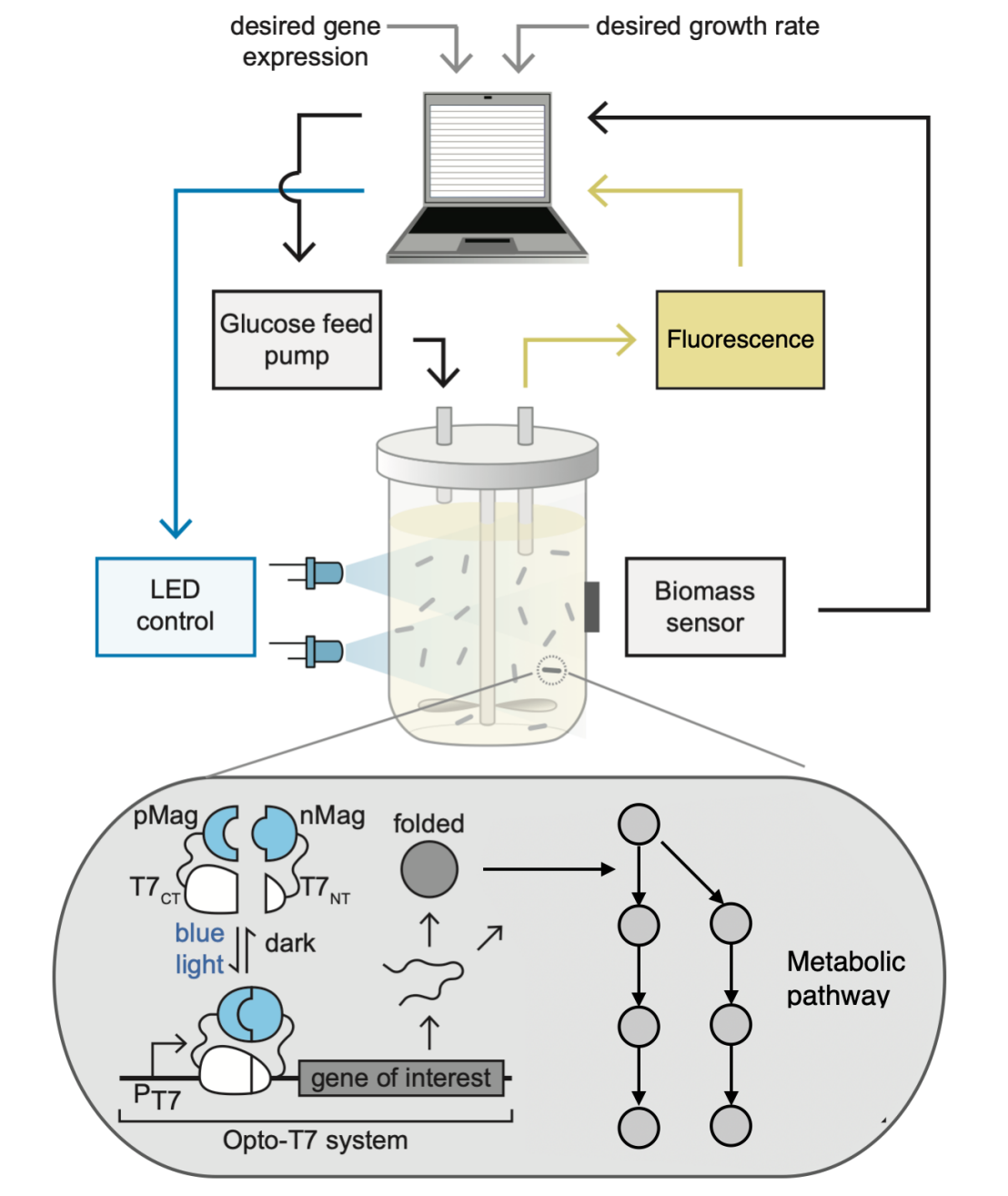
One way to realize cybergenetic control is by interfacing the living cells with a digital computer which implements the control system. The interface is achieved using light (optogenetics) or chemical inducers. In the case of light, the cells are genetically engineered to host a light responsive circuits which activates a biological process of interest, e.g. gene expression. The output of this biological process, e.g. protein concentration, is sensed in real time, typically by measuring flurorescene intensity, and the measured values are fed to the computer. The computer implements a control system whose commands are translated into light signals that are delivered to the cells through an LED, thereby closing the feedback loop. This in silico control configuration enables flexible and fast implmentation of sophisticated control systems that can be rapidly modified and optimized.
High Performance Optogenetic Regulators
OptoT7 optogenetic system for light-induced orthogonal transcription in bacteria
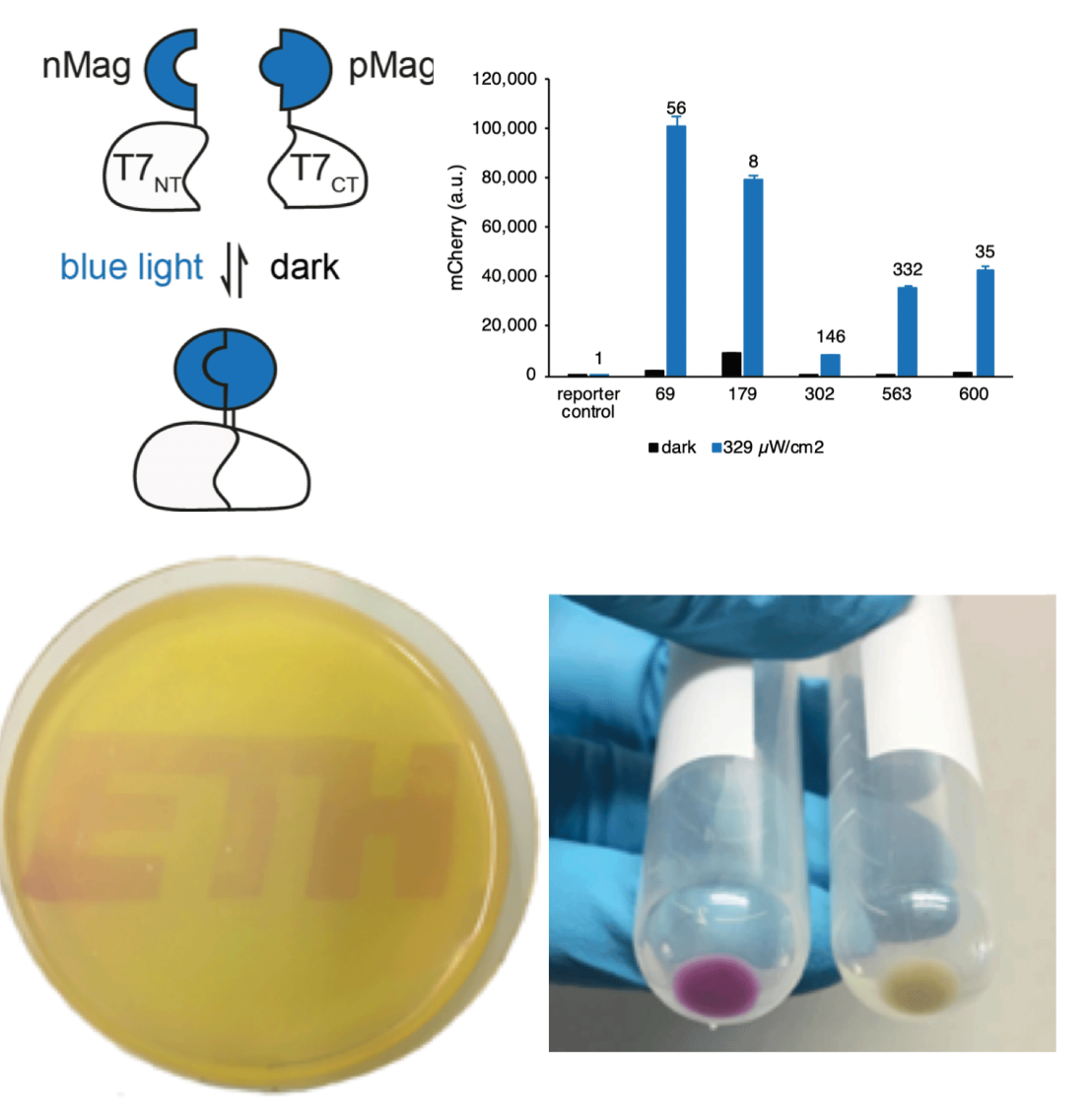
The limited toolset for light control in bacteria motivated us to develop a light-inducible transcription system that is independent from cellular regulation through the use of an orthogonal RNA polymerase. We engineered blue light-responsive T7 RNA polymerases (Opto-T7RNAPs) that show properties such as low leakiness of gene expression in the dark state, high expression strength when induced with blue light with an inducible range of more than 300-fold, while returning to inactive state within mintues in the dark. Our approach expands the applicability of using light as an inducer for gene expression independent from cellular regulation and allows for a more reliable dynamic control of synthetic and natural gene networks.
EL222 optogenetic system for light-induced transcription in yeast
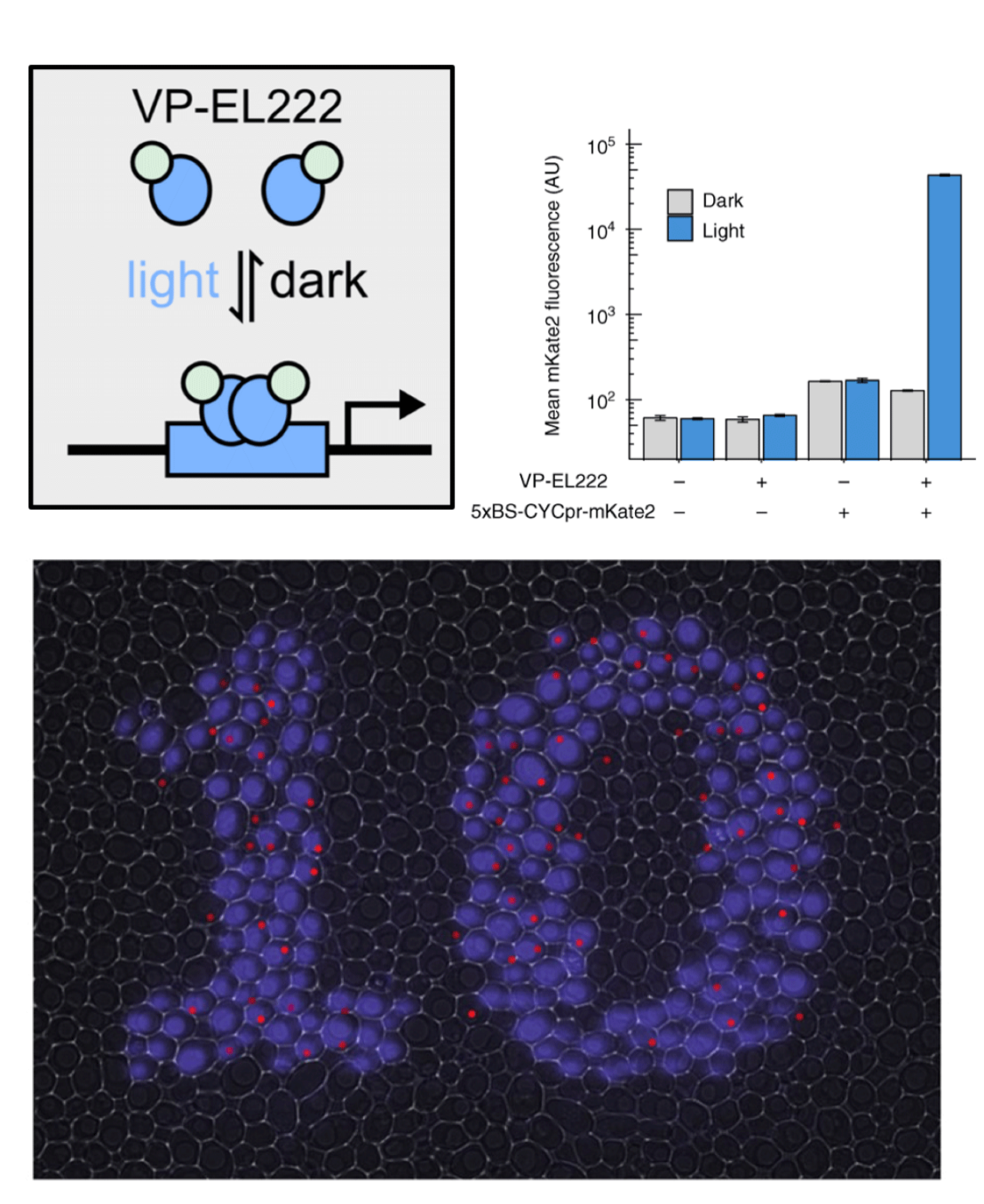
To meet the speed and reversibility requirements of control applications, we employed a photosensitive transcription factor consisting of a nuclear localization signal, the VP16 transactivation domain, and the bacterially derived LOV-domain protein EL222 to build a blue light transcriptional activation system. Light timulation induces structural changes in EL222 in seconds, leading to transcriptional activation, while deactivating within 1 min in the dark. This system is at the heart of multiple optogenetic applications in yeast, in our lab and other labs.
Robust and Precise Control of Cell Populations
Automated optogenetic feedback control of gene expression
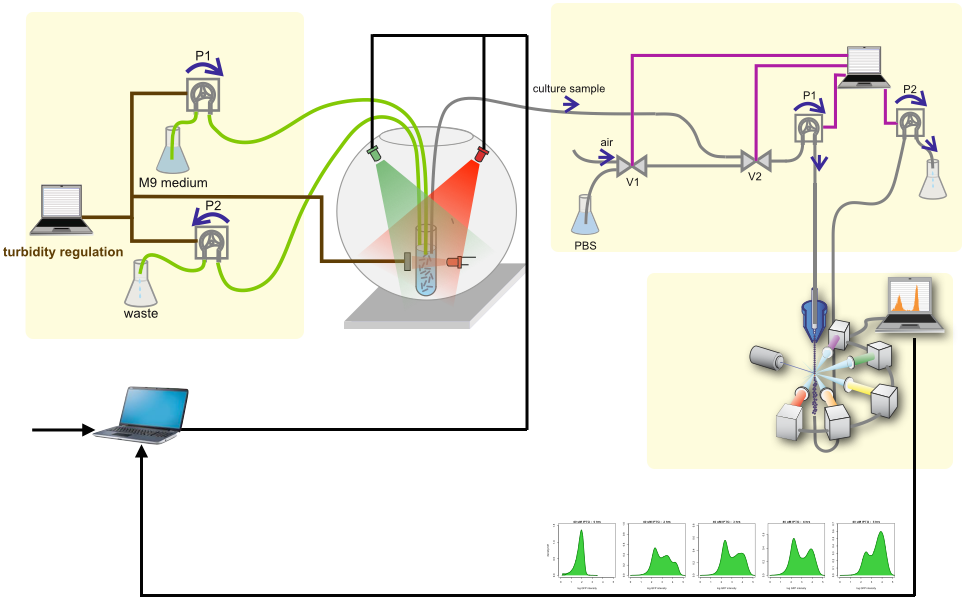
Dynamic control of gene expression can have far-reaching implications for biotechnological applications and biological discovery. Thanks to the advantages of light, optogenetics has emerged as an ideal technology for this task. We deveoped a novel fully automatic experimental platform for the robust and precise long-term optogenetic regulation of protein production in liquid Escherichia coli cultures. Using a computer-controlled light-responsive system we accurately tracked prescribed dynamic green fluorescent protein expression profiles through the application of feedback control, and showed that the system adapts to global perturbations such as nutrient and temperature changes. We demonstrated the efficacy and potential utility of our approach by placing a key metabolic enzyme under optogenetic control, thus enabling dynamic regulation of the culture growth rate with potential applications in bacterial physiology studies and biotechnology.
In silico feedback for in vivo regulation of a gene expression circuit
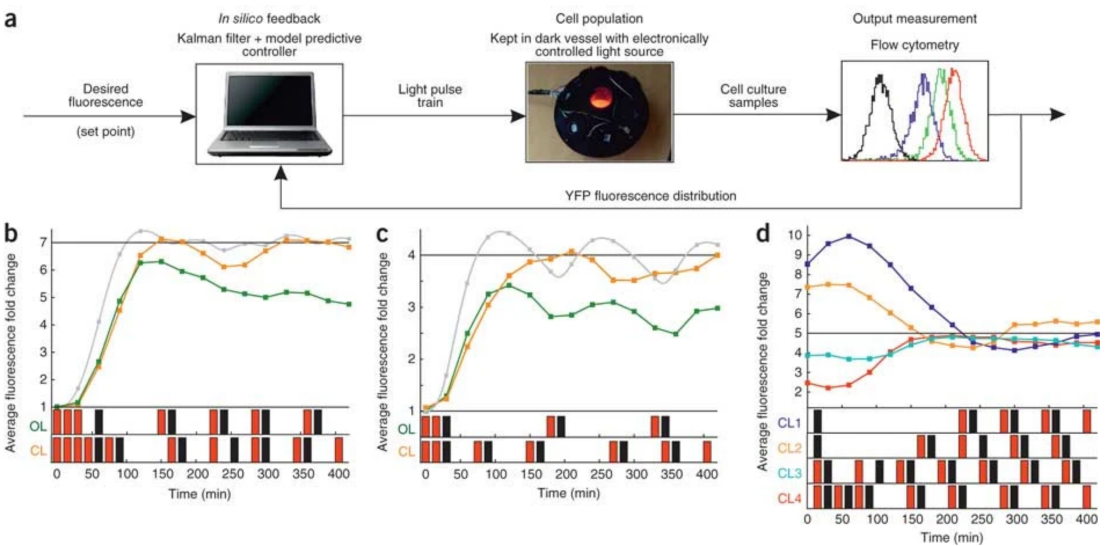
Working with collaborators from the Lygeros and El-Samad groups, we showed that difficulties in regulating cellular behavior with synthetic biological circuits may be circumvented using in silico feedback control. By tracking a circuit's output in Saccharomyces cerevisiae in real time, we precisely controled its behavior using an in silico feedback algorithm to compute regulatory inputs implemented through a genetically encoded light-responsive module. This was the first time a computer was interfaced with living cells to control gene expression.
Scaling up: optogenetic bioreactors
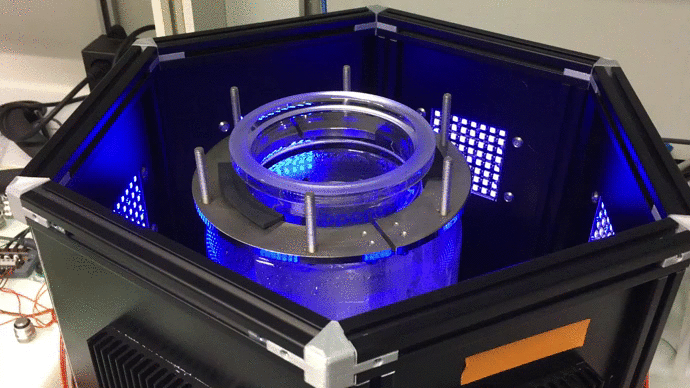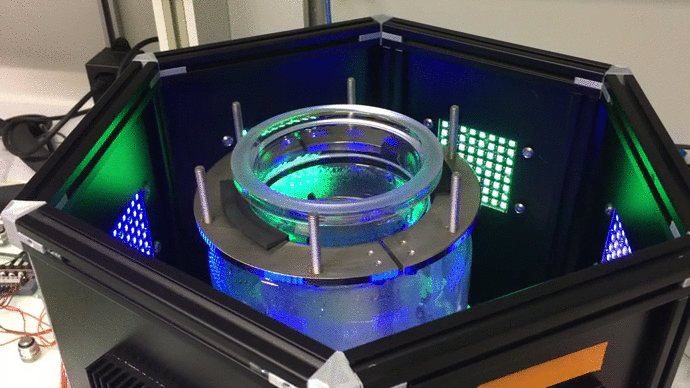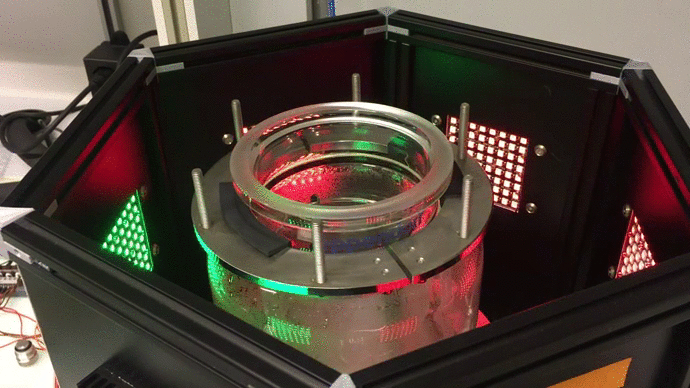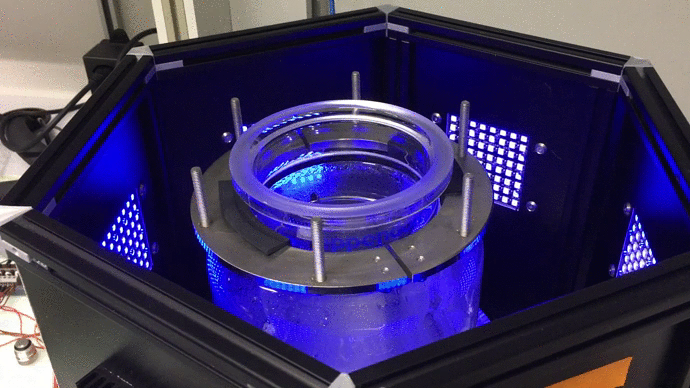
We have designed and built new bioreactors outfitted with optogenetic actuators and various sensors. With this platform, we are applying cybergenetic population control for enhacing product yield in yeast and E. coli at larger scales.
Dirk Benzinger and Mustafa Khammash. "Pulsatile inputs achieve tunable attenuation of gene expression variability and graded multi-gene regulation," Nature Communications, vol. 9, pp. 3521, London: Nature Publishing Group, 2018.external page DOI: 10.1038/s41467-018-05882-2
Armin Baumschlager, Stephanie K. Aoki and Mustafa Khammash. "Dynamic Blue Light-Inducible T7 RNA Polymerases (Opto-T7RNAPs) for Precise Spatiotemporal Gene Expression Control," ACS Synthetic Biology, 6 (11): 2157-2167, London: American Chemical Society, 2017. external page doi:10.1021/acssynbio.7b00169
A. Milias-Argeitis, M. Rullan, S. Aoki, P. Buchmann, and M. Khammash. "Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth," Nature Communications, 2016. external page doi:10.1038/ncomms12546
Andreas Milias-Argeitis, Stefan Engblom, Pavol Bauer and Mustafa Khammash. "Stochastic focusing coupled with negative feedback enables robust regulation in biochemical reaction networks," Interface: Journal of the Royal Society, vol. 12: no. 113, pp. 20150831, London: Royal Society, 2015. external page DOI: 10.1098/rsif.2015.0831
J. Ruess, F. Parise, A. Milias-Argeitis, M. Khammash, and J. Lygeros. "Iterative experiment design guides the characterization of a light-inducible gene expression circuit," Proceedings of the National Academy of Sciences, 2015. external page doi:10.1073/pnas.1423947112
A. Gupta, C. Briat, and M. Khammash, "A Scalable Computational Framework for Establishing Long-Term Behavior of Stochastic Reaction Networks," PLoS Computational Biology, 2014. external page doi:http://dx.doi.org/10.1371/journal.pcbi.1003669
C. Briat and M. Khammash, "Integral population control of a quadratic dimerization process," IEEE Conference on Decision and Control, 2013, pp. 3367-3372. external page doi:10.1109/CDC.2013.6760398
C. Briat and M. Khammash, "Computer control of gene expression: Robust setpoint tracking of protein mean and variance using integral feedback," IEEE Conference on Decision and Control, 2012, pp. 3582-3588. external page doi:10.1109/CDC.2012.6426720
A. Milias-Argeitis, S. Summers, J. Stewart-Ornstein, I. Zuleta, D. Pincus, H. El-Samad, M. Khammash, and J. Lygeros. "In silico feedback for in vivo regulation of a gene expression circuit," Nature Biotechnology, 2011. external page doi:10.1038/nbt.2018