Computer Control of Single Cells
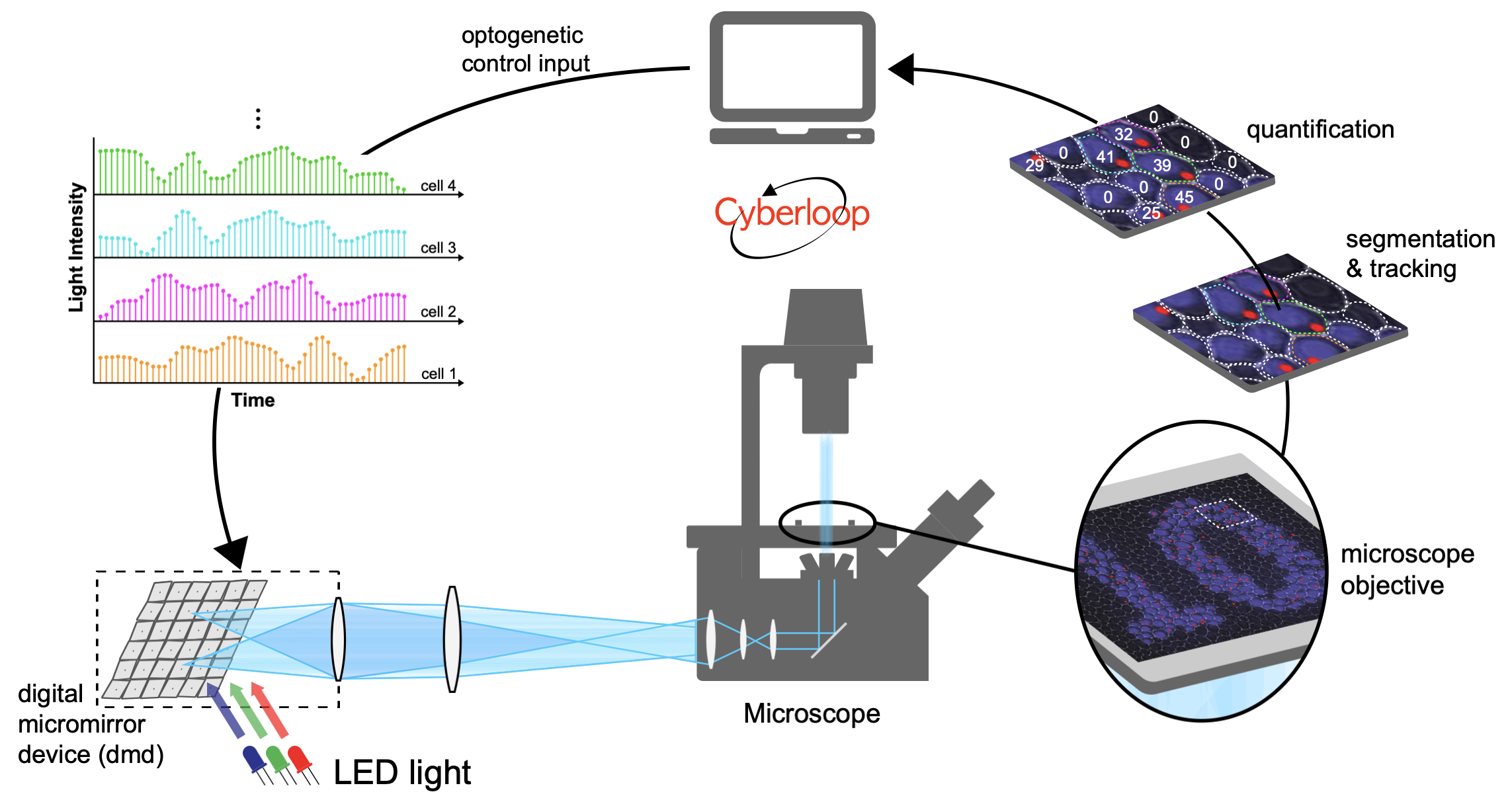
Feedback Control of Transcription in Single Cells
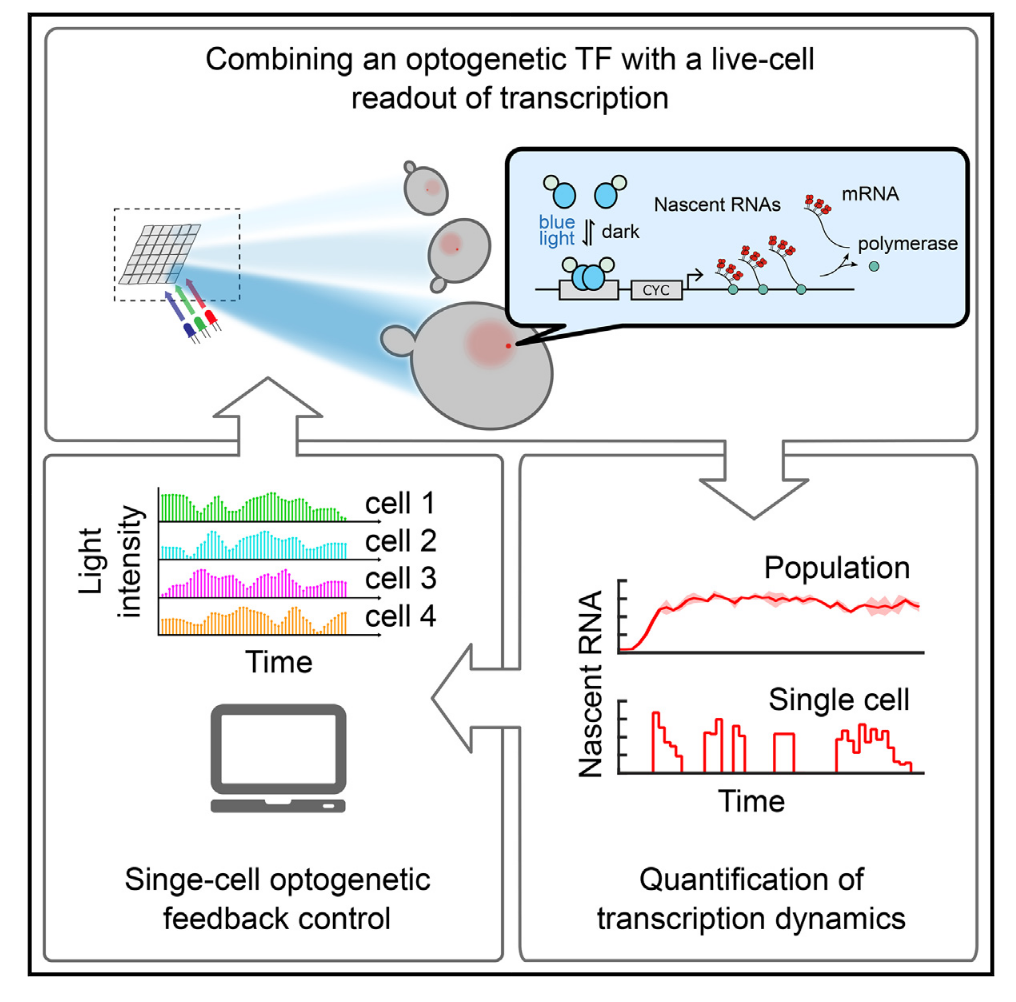
Transcription is a highly regulated and inherently stochastic process. The complexity of signal transduction and gene regulation makes it challenging to analyze how the dynamic activity of transcriptional regulators affects stochastic transcription. By combining a fast-acting, photo-regulatable transcription factor with nascent RNA quantification inlive cells and an experimental setup for precise spatiotemporal delivery of light inputs, we constructed a platform for the real-time, single-cell interrogation of transcription in Saccharomyces cerevisiae. We showed that transcriptional activation and deactivation are fast and memoryless. By analyzing the temporal activity of individual cells, we found that transcription occurs in bursts, whose duration and timing are modulated by transcription factor activity. Using our platform, we regulated transcription via light-driven feedback loops at the single-cell level. Feedback markedly reduced cell-to-cell variability and led to qualitative differences in cellular transcriptional dynamics. Our platform establishes a flexible method for studying transcriptional dynamics in single cells.
Using the Cyberloop for studying pattern formation through emulated cell-to-cell signalling
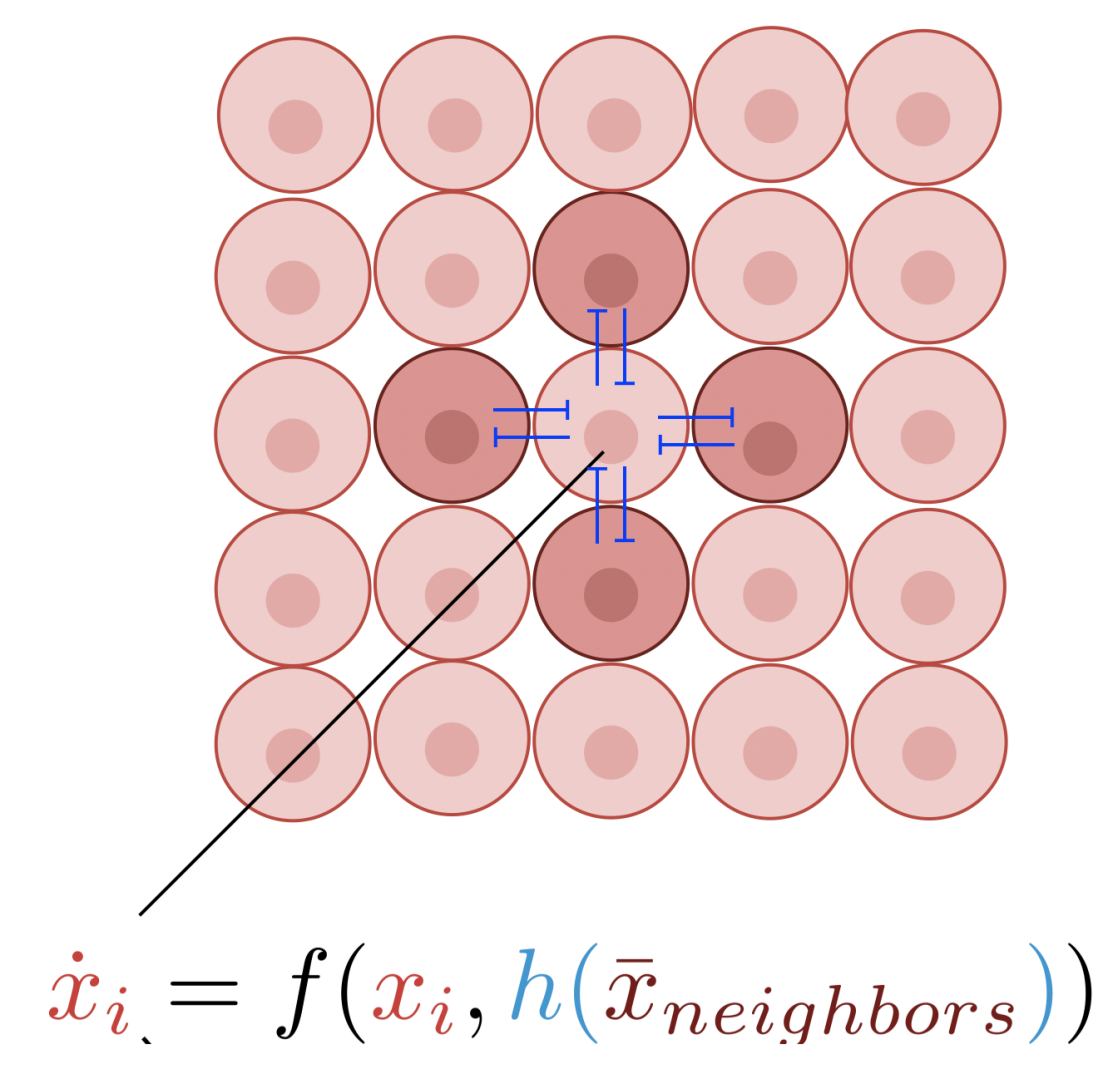
Designing and implementing synthetic biological pattern formation remains challenging due to underlying theoretical complexity as well as the difficulty of engineering multicellular networks biochemically. Here, we used our Cyberloop platform to introduce a cell-in-the-loop approach where living cells interact through in silico signaling, establishing a new testbed to interrogate theoretical principles. We presented an easy-to-use theoretical test to predict the emergence of contrasting patterns in gene expression among laterally inhibiting cells. Guided by the theory, we experimentally demonstrated spontaneous checkerboard patterning where cell-to-cell signaling is emulated with light inputs calculated in silico from real-time gene expression measurements. The scheme successfully produced spontaneous, persistent checkerboard patterns for systems of sixteen patches, in quantitative agreement with theoretical predictions. Our research illustrates the potential of our Cyberloop for understanding pattern formation through emulated cell-to-cell communication.
Understanding the spatiotemporal signaling dynamics prior and during gastrulation
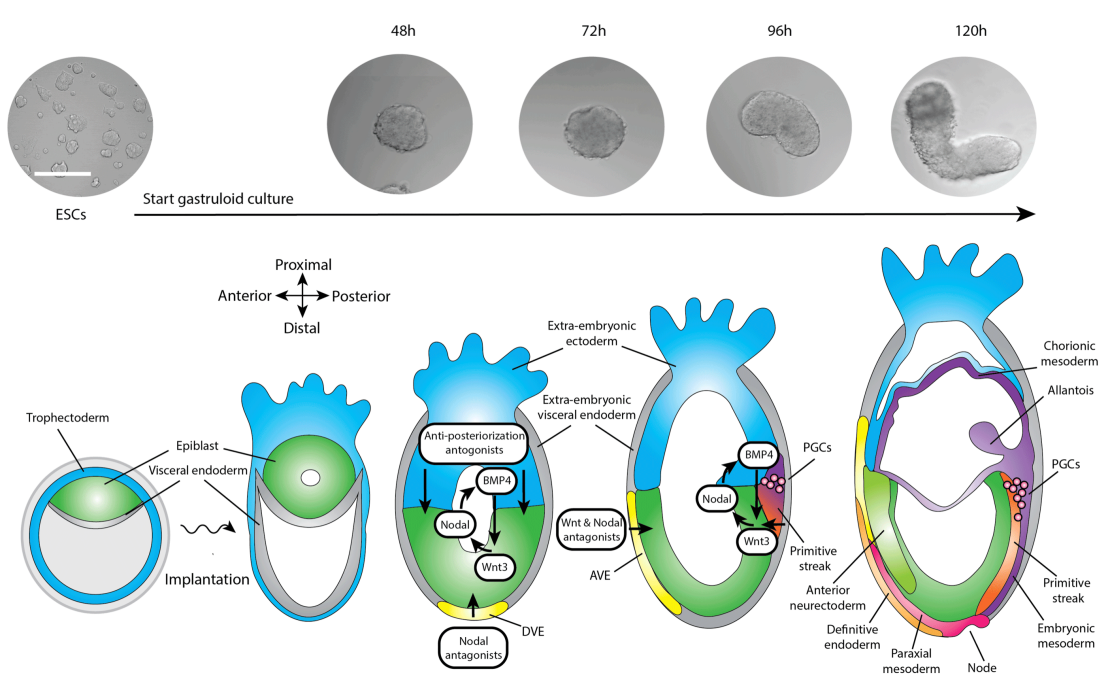
As soon as the mammalian embryo inplants in the uterus, gastrulation will start in the epiblast. During gastrulation, the embryonic axes as well as the primary three germ layers are formed: endoderm, mesoderm and ectoderm. Signaling from the extra-embryonic tissues is essential for the onset of gastrulation. However, due to the inaccessibility of the mammalian embryo at this stage of development, it remains unknown what the spatiotemporal dynamics are of these signaling factors during gastrulation and how these affect each other.
We are using our Cyberloop to study signaling dynamics during gastrulation in an organoid model for early embryonic development: gastruloids. Currently, gastruloids resemble many features of early developing embryos but lack a clear internal organisation and a full anterior axis probably due to the absence of extra-embryonic tissues. Our platform enables the simulation of signaling from extrambryonic tissues using optogenetics to get insight into the signaling dynamics contributing to gastrulation. This research is aimed at establishing of an embryonic organoid model that more faithfully recapitulates in vivo embryonic development.
Melinda Liu Perkins, Dirk Benzinger, Murat Arcak and Mustafa Khammash. "Cell-in-the-loop pattern formation with optogenetically emulated cell-to-cell signaling," Nature Communications, vol. 11: no. 1, pp. 1355, London: Nature Publishing Group, 2020.
external page DOI: 10.1038/s41467-020-15166-3
Dirk Benzinger, Marc Rullan, Andreas Milias-Argeitis and Mustafa Khammash. "Studying and Regulating Transcription Using Single-Cell Optogenetics: Principles of Systems Biology," No. 30. Cell Systems, vol. 7: no. 1, pp. 1-1, Cambridge: Cell Press, 2018. external page Article
Marc Rullan, Dirk Benzinger, Gregor W. Schmidt, Andreas Milias-Argeitis, Mustafa Khammash. "An Optogenetic Platform for Real-Time, Single-Cell Interrogation of Stochastic Transcriptional Regulation," Molecular Cell, 2018. external page Article
Armin Baumschlager, Stephanie K. Aoki and Mustafa Khammash. "Dynamic Blue Light-Inducible T7 RNA Polymerases (Opto-T7RNAPs) for Precise Spatiotemporal Gene Expression Control," ACS Synthetic Biology, 6 (11): 2157-2167, London: American Chemical Society, 2017. external page doi:10.1021/acssynbio.7b00169