Research
▪ Molecular control of (stem) cell fate decisions
▪ Continuous long-term single-cell quantification
▪ Blood, pluripotent, bone and cancer (stem) cells
Many organs like blood, skin and intestine have the amazing capability for life-long regeneration and repair. Every second of our life, millions of the right type of cells have to be produced in these tissues at the right time and place.
Fates of all individual cells underlie tissue health and disease.
We analyze how these cell fates are controlled at the molecular level.
This will allow manipulation of cell behavior for clinical therapy. Due to their huge therapeutic potential in regenerative medicine, stem and progenitor cells are of particular interest.
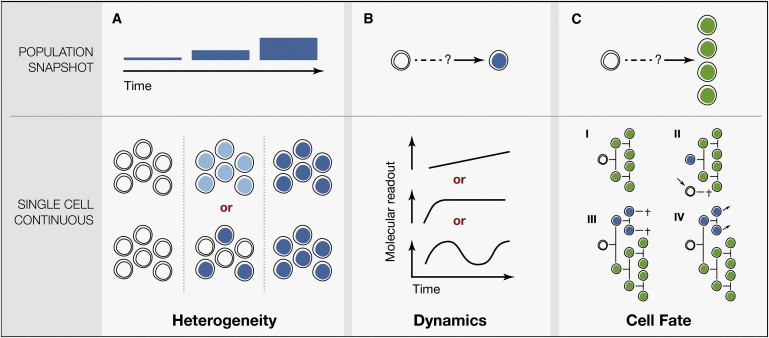
Although usually ignored, the ability to quantify cell fate decisions and molecular dynamics continuously and at the single cell level is a crucial prerequisite when trying to understand molecular cell fate control.
Snapshot analyses with data from only few time points usually allow too many different interpretations about underlying mechanisms. This is the reason for many long standing scientific disputes.
Selected Reviews
Kull, Journal of Experimental Medicine 2021
Loeffler, Blood 2019
Skylaki, Nature Biotechnology 2016
Etzrodt, Cell Stem Cell 2014
Hoppe, Nature Cell Biology 2014
Schroeder, Nature Methods 2011
Schroeder, Nature 2008
Schroeder, Annals - New York Academy of Science 2005
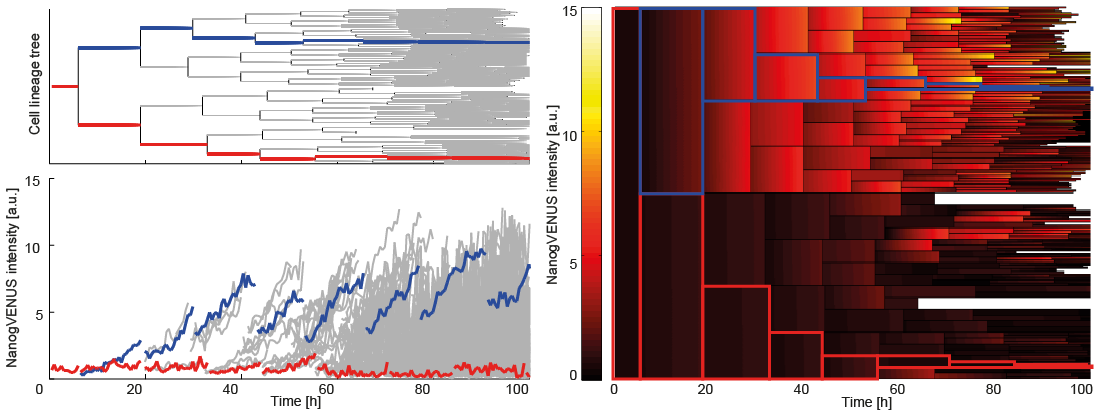
For a better understanding of the cellular and molecular dynamics underlying health and disease, we therefore develop bioimaging approaches to allow the long-term quantification of cellular and molecular behavior of all individual cells in cell cultures for up to several weeks.
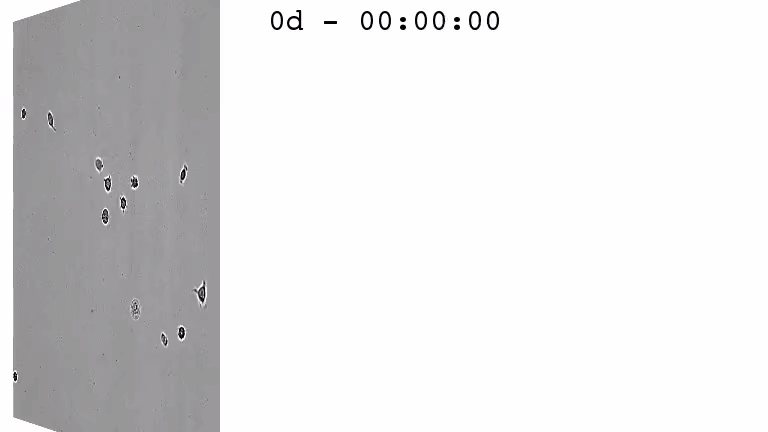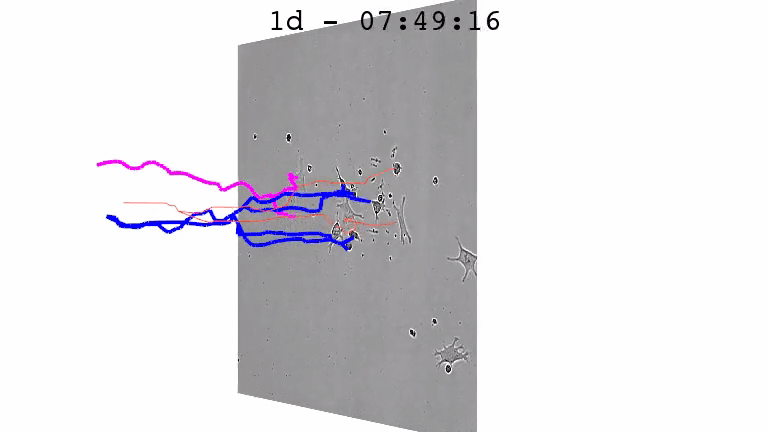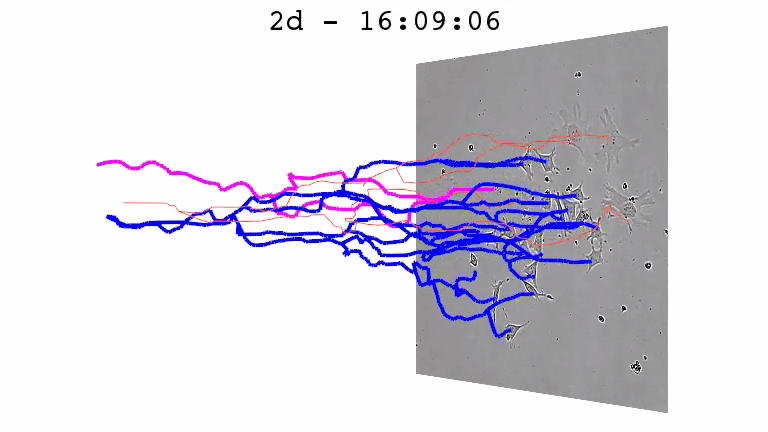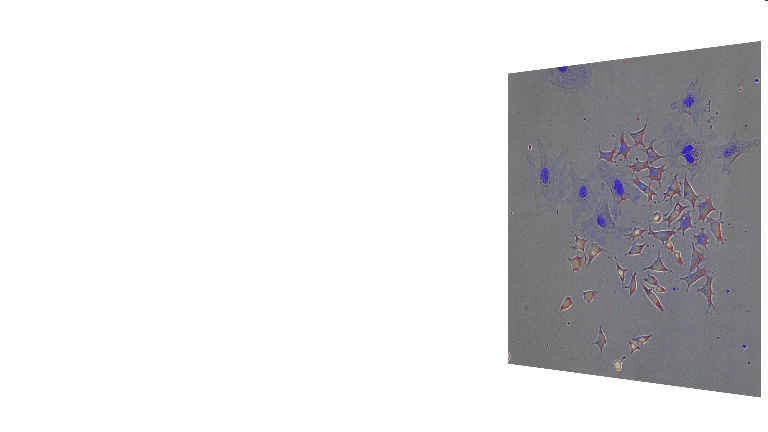
We combine
- (stem) cell purification and culture
- molecular manipulation
- microscopic imaging
- software development for image acquisition and analysis
- engineering of experimental hardware
to enable the documentation and analysis of cell fate decisions and their molecular control with improved resolution.

This combination of novel tools from different disciplines now allows much improved identification of the exact role of cell-intrinsic molecules, signals from the microenvironment and synthetic compounds on (stem) cell behavior.
Selected Publications
Wehling, Blood 2022
Kull, Blood 2022
Loeffler, Nature 2019
Loeffler, Blood 2018
Hilsenbeck, Bioinformatics 2017
Buggenthin, Nature Methods 2017
Hilsenbeck, Nature Biotechnology 2016
Skylaki, Nature Biotechnology 2016
Hoppe, Nature 2016
Kokkaliaris, Blood 2016
Filipczyk, Nature Cell Biology 2015
Eilken, Nature 2009
Rieger, Science 2009
Development of custom software has proven crucial for every step of data acquisition, curation and analysis.
Published software can be downloaded here
Selected Publications
Wehling, Blood 2022
Kunz, Cell Stem Cell 2019
Coutu, Nature Methods 2017
Hilsenbeck, Bioinformatics 2017
Buggenthin, Nature Methods 2017
Hilsenbeck, Nature Biotechnology 2016
Skylaki, Nature Biotechnology 2016
We design and produce custom hardware for improved laboratory experimentation, culture, manipulation and retrieval of cells and imaging. Products include microfluidic chips, larger 3D-printed devices, small robots and electronics - all including their required control software.

Published hardware can be downloaded here
Selected Publications
Dettinger, BioRx 2021 / Nature Communications 2022
Dettinger, Lab on a Chip 2020
Dettinger, Analytical Chemistry 2018
We analyze the function and regulation of:
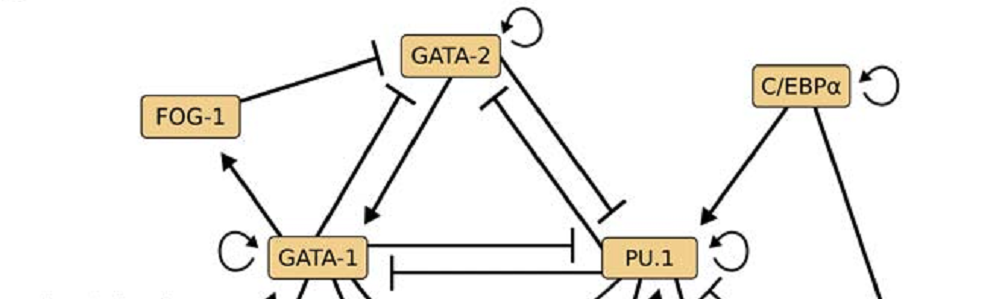
Transcription factors and their networks control cell fates. We therefore develop approaches to manipulate and quantify components, topology, wiring and dynamics of transcription factor networks - ideally live in single cells.
Selected Publications
Ahmed, Journal of Experimental Medicine 2022
Ahmed, Stem Cell Reports 2020
Etzrodt, Blood 2019
Hoppe, Nature 2016
Filipczyk, Cell Stem Cell 2013
Filipczyk, Nature Cell Biology 2015
Krumsiek, PLoS One 2011
Schroeder, J Immunol 2003, PNAS 2003, EMBO Journal 2000
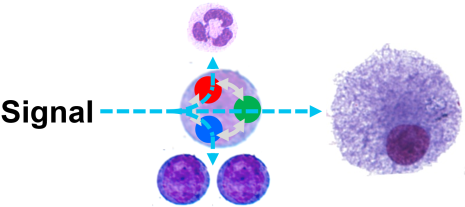
We analyze the influence of signaling pathways and their networks in directing cell fate choices through transcription factors by quantitative long-term live single-cell bioimaging.
Selected Publications
Kull, Blood 2022
Wang, Blood 2021
Etzrodt, Blood 2019
Hastreiter, Stem Cell Reports 2018
Endele, Blood 2017
Kokkaliaris, Blood 2016
Endele, Exp Cell Res 2014
Kimura, Blood 2009
Rieger, Science 2009
Schroeder, Mech Dev 2006, J Immunol 2003, EMBO Journal 2000
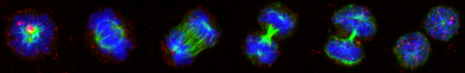
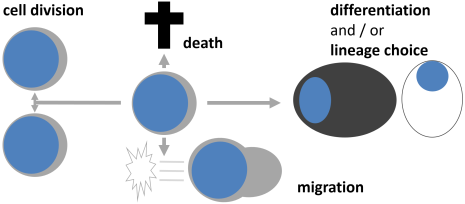
Continuous single cell quantification is crucial for the analysis of asymmetric cell division. We analyze its role and regulation in hematopoietic and pluripotent stem and progenitor cells.
Selected Publications
Wehling, Blood 2022
Loeffler, Blood 2022
Loeffler, Current Opinion in Hematology 2021
Loeffler, Nature 2019
Loeffler, Annals - New York Academy of Science 2019
Schroeder, Cell Stem Cell 2007
We work in these human and mouse cell systems:
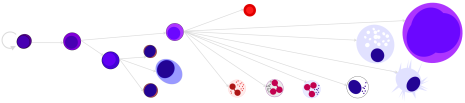
Selected Publications
Wehling, Blood 2022
Kull, Blood 2022
Ahmed, Journal of Experimental Medicine 2022
Loeffler, Blood 2022
Wang, Blood 2021
Kokkaliaris, Blood 2020
Ahmed, Stem Cell Reports 2020
Loeffler, Nature 2019
Etzrodt, Blood 2019
Buggenthin, Nature Methods 2017
Endele, Blood 2017
Hoppe, Nature 2016
Kokkaliaris, Blood 2016
Schroeder, Cell Stem Cell 2010
Eilken, Nature 2009
Kimura, Blood 2009
Rieger, Science 2009
Schroeder, EMBO Journal 2000

Selected Publications
Filipczyk et al., Nature Cell Biology 2015
Filipczyk et al., Cell Stem Cell 2013
Warlich et al., Mol Ther 2011
Eilken et al., Nature 2009
Schroeder et al., Mech Dev 2006
Schroeder et al., PNAS 2003
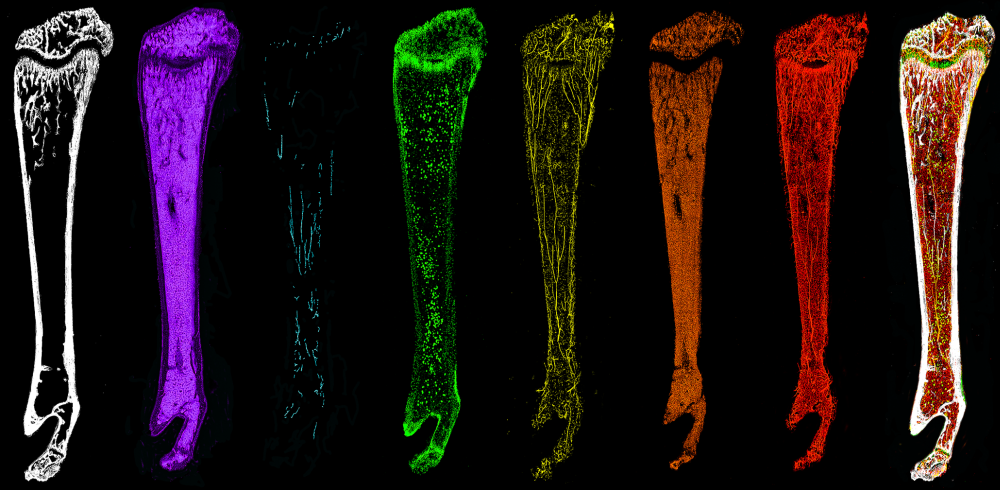
Skeletal or mesenchymal stem cells show great promise for therapy but their biology remains poorly understood. To better characterize these cells, and their role as a niche for other cell types in the bone marrow, we are using novel approaches for multidimensional FACS analyses, lineage tracing approaches and multicolor confocal whole bone imaging.
Selected Publications
Kokkaliaris et al., Blood 2020
Kunz et al., Cell Stem Cell 2019
Bourgine et al., PNAS 2018
Bourgine et al., Cell Stem Cell 2018
Coutu et al., Nature Methods 2018
Coutu et al., Nature Biotechnology 2017