alerT

Automated Live Event Recognition in Time lapse data.
alerT helps you to identify events of interest (e.g. specific cellular or molecular behaviors) and find their physical location in time-lapse imaging experiments in near real-time.
Overview
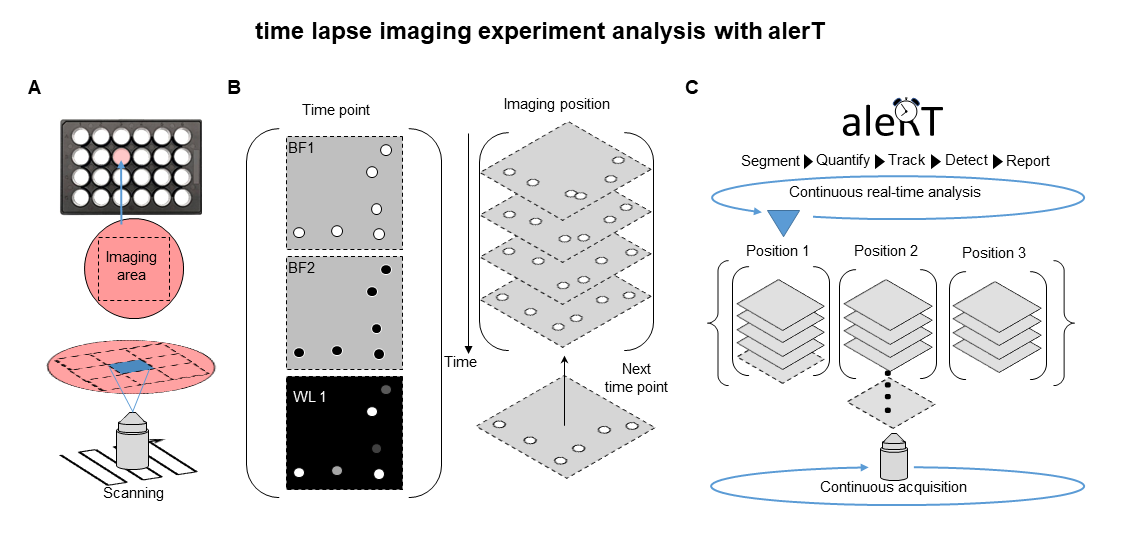
During time lapse imaging experiments, automated imaging iterates through user-defined positions and acquires images over time (Fig 1A). Acquired images (possibly in different imaging configurations / channels) are bundled per time point and position (Fig 1B).
alerT monitors the experiment in real time to recognize incoming new time points, and segments, quantifies and tracks cells to then detect and report user-defined molecular or cellular dynamics (Fig. 1C).
Input structure
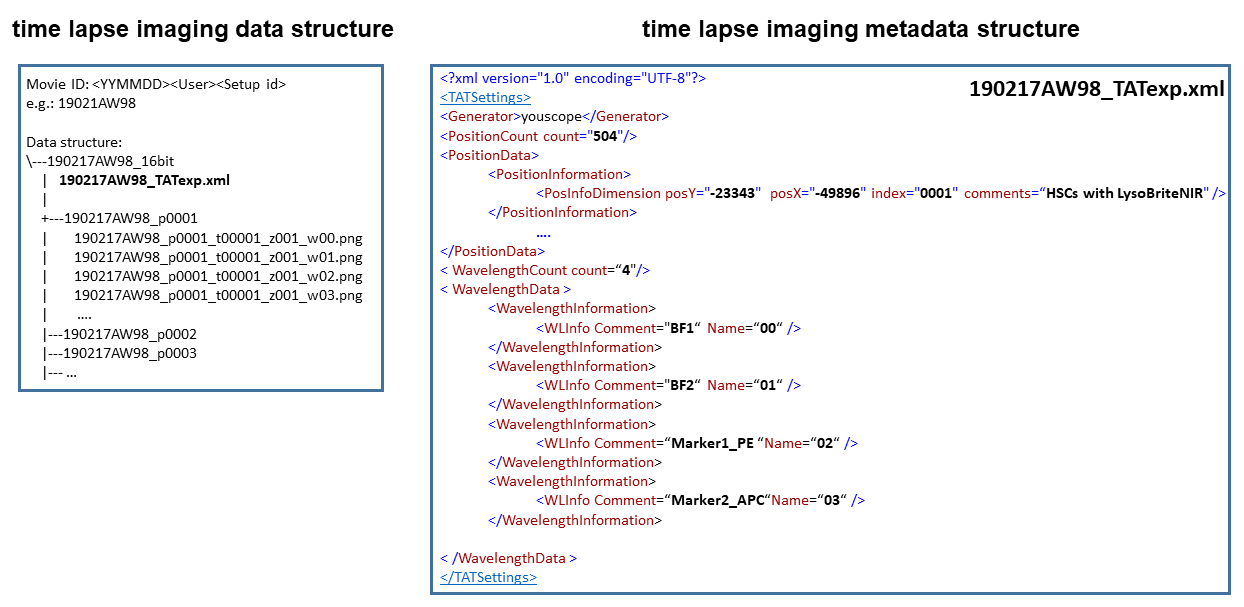
alerT requires the time lapse imaging experiment (“movie”) data to be in the following fixed folder, data and naming structure (Fig. 2):
Top level directory name: <Movie ID>_16bit
The movie ID is the prefix for all subdirectories and their contained files. It is constructed as follows:
<YYMMDD><User Initials><Microscope ID>
(e.g. 201125AW15 for an experiment started on 25 November 2020 by Arne Wehling on microscope setup #15)
The top level directory name in this example would be: 201125AW15_16bit
Imaging positon directory names:
<Movie ID>_pXXXX
pXXXX: position number (in four digits, 0001 - 9999)
(e.g. 201125AW15_p0001 for the first position in movie 201125AW15)
alerT expects that images are stored as single color 16-bit (bit depth) PNGs per position, time point, and channel in the position-specific subdirectories.
Image file names:
<Movie ID>_pXXXX_tXXXXX_z001_wXX.png
tXXXXX: acquisition time points (in 5 digits, 00001-99999). These are discrete time points. Their absolute time (date) is stored in the metadata file(s) as described below.
wXX: channel number (in 2 digits, 00-99). This is an arbitrary number for e.g. brightfield or fluorescence channels. We use 00 for a channel that is acquired at all positions at every timepoint (typically used for cell tracking). The individual hardware settings of different channels are stored in the metadata file(s) as described below. For additional channels, the_wXX suffix increases by +1. If there were three channels acquired in the movie, wXX is w00, w01 and w02.
(e.g. 201125AW15_p0001_t00001_z001_w00.png for the first image in position 1 of channel 0 in movie 201125AW15)
IMPORTANT:
For segmentation, alerT currently expects 2 brightfield images, named BF1 and BF2, to be acquired per time point and position and to be described within the metadata.
alerT currently also expects 2 more images (e.g. fluorescent images) that must carry the suffixes APC and PE in the metadata (see Fig. 2 time lapse imaging metadata structure).
alerT currently thus expects 4 images per time point. E.g. for the first time point in the first positon in movie 201125AW15:
· 201125AW15_p0001_t00001_z001_w00.png
· 201125AW15_p0001_t00001_z001_w01.png
· 201125AW15_p0001_t00001_z001_w02.png
· 201125AW15_p0001_t00001_z001_w03.png
To function properly, the different channels (w00 – w03) must be named BF1, BF2, _PE and _APC within the WavelengthData field of the metadata file as explained in the next section. The order is not important, e.g. _PE does not need to be w02, but must be consistent throughout all positions and time points of the movie.
Metadata structure
Metadata is stored in a required XML file named in <Movie ID>_TATexp.xml (e.g. 201125AW15_TATexp.xml).
The file’s header and required fields are specified in Fig. 3A. You must mimic this file to communicate meta information about the time lapse imaging experiment to alerT.
The field PositionCount tells alerT how many positions to expect listed under PositionData. Each position lives within its own PositionInformation field, with PositionInfoDimension carrying crucial information such as X/Y position (important for plotting event reports) and the position’s index and imaging and culture condition.
The field WavelengthCount tells alerT how many different channels have been acquired, with their names and sequence specified in separate WavelengtInformation fields within the WavelengthData field.
alerT algorithm process and output
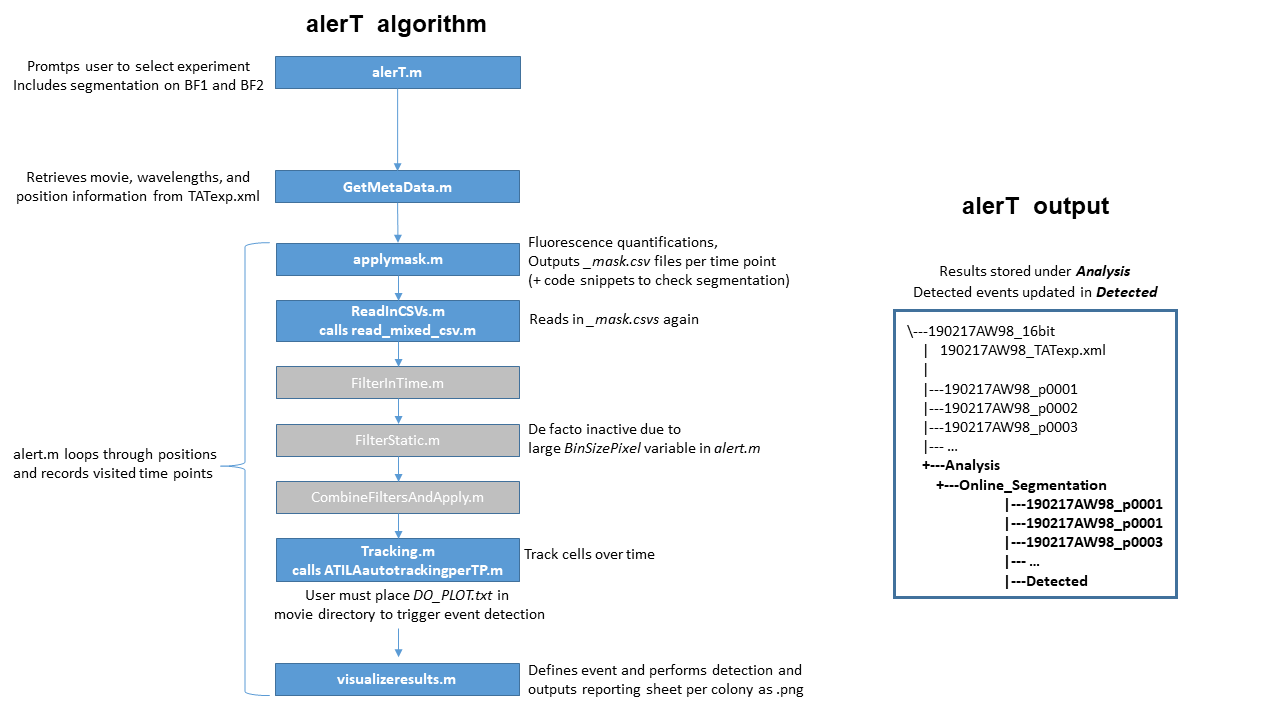
Executing the alert.m script will prompt the user to specify the ongoing time lapse imaging experiment’s top-level directory.
alerT then reads the timelapse imaging experiment metadata from the _TATexp.xml file using the GetMetaData.m script.
Segmentation per time point occurs within the alert.m script, and applymask.m carries out quantifications and corrections (Fig. 3 alerT algorithm) and writes results to CSV files under Analysis > Online_Segmentation within the time lapse imaging experiment’s top-level directory. Herein, the position-wise folder structure is replicated (Fig. 3 alerT output).
To perform tracking, alerT uses its CSV output in its Tracking.m script. Tracking is performed in a sliding time window approach that can halt per position and move to the next to wait for newly incoming time points to guarantee continuous quasi real-time quantification and tracking. The tracking algorithm itself is wrapped within the function ATILAautotrackingperTP.m, whose functionality is explained in Chapter 1 of Arne Wehling's PhD thesis.
Event detection is customizable and currently returns all colonies in their 2-cell state (i.e. having undergone one division) to the user. Detected events are constantly updated and saved to the Detected folder under the Analysis > Online_Segmentation directory.
IMPORTANT: The event definition and reporting script visualizeresults.m only becomes active after placing an empty file named DO_PLOT.txt into the time lapse imaging experiment’s top-level directory.
If you want to change visualizations, event detection or reporting, please modify this script to your needs.
Code
Download alerT.zip (ZIP, 35 KB)
36 KB - contains the following scripts:
alerT.m
applymask.m
ATILAautotrackingperTP.m
CombineFiltersAndApply.m
FilterInTime.m
FilterStatic.m
GetMetaData.m
ReadInCSVs.m
read_mixed_csv.m
subplottight.m
Tracking.m
visualizeresults.m
Download and unpack the zip file.
Add the resulting alerT directory to your MATLAB for execution.
Reference
If you use alerT, please cite the following paper:
Wehling, Loeffler, Zhang, Kull, Donato, Szczerba, Camargo Ortega, Lee, Moor, Goettgens, Aceto and Schroeder
external page Combining single-cell tracking and omics improves blood stem cell fate regulator identification
Blood 2022, 10.1182/blood.2022016880
License
alerT is free software: you can redistribute it and/or modify it under the terms of the 3-Clause BSD License.
This software is provided by the copyright holders and contributors “AS IS” and any EXPRESS or IMPLIED WARRANTIES, including, but not limited to, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. See the BSD-3-License for more details (external page https://opensource.org/licenses/BSD-3-Clause).