High-throughput experimenting
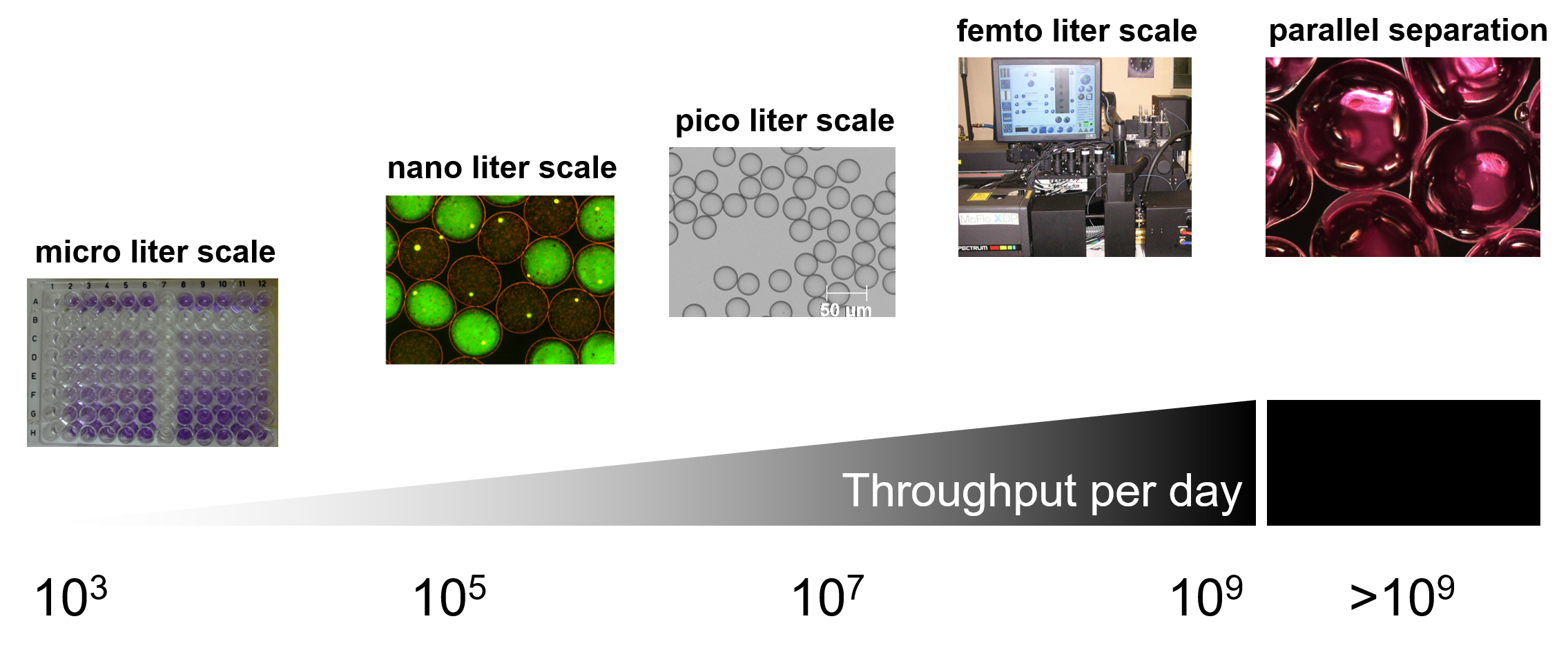
High Throughput Experimenting (HTE) refers to the massive parallelization of experiments. Specifically, we apply rapid large-scale analysis of either proteins, multiple component cascades or even whole bacterial cells. The throughput is usually determined by the reaction volume, which ranges from microliter (low throughput) to femtoliter (high throughput) scale, or by the separation principle.
In our case the reaction parameters of a system or – even more importantly – the genetic starting material for a protein (system) or a cell are systematically or randomly varied. They are then read out via a specific phenotype and investigated at the same time in order to identify interesting study objects (screening) or even to gather insights into the dynamics for the entire population. HTE is a technique that allows the execution of large numbers of experiments in parallel while requiring less effort per experiment when compared to traditional means of experimentation. HTE is becoming standard practice in biotechnology, biology and pharmaceutical laboratories across the world.

One of the most common and standard reaction scales in our lab is the microliter range, usually in the form of microtiter plates. If multiple parameters of a specific reaction need to be addressed simultaneously, these plates are no longer handled by individuals but by increasingly sophisticated robots at the D-BSSE lab automation facility. With this powerful installation a huge space of experimental variations (up to 103 conditions) can be easily covered in a day.
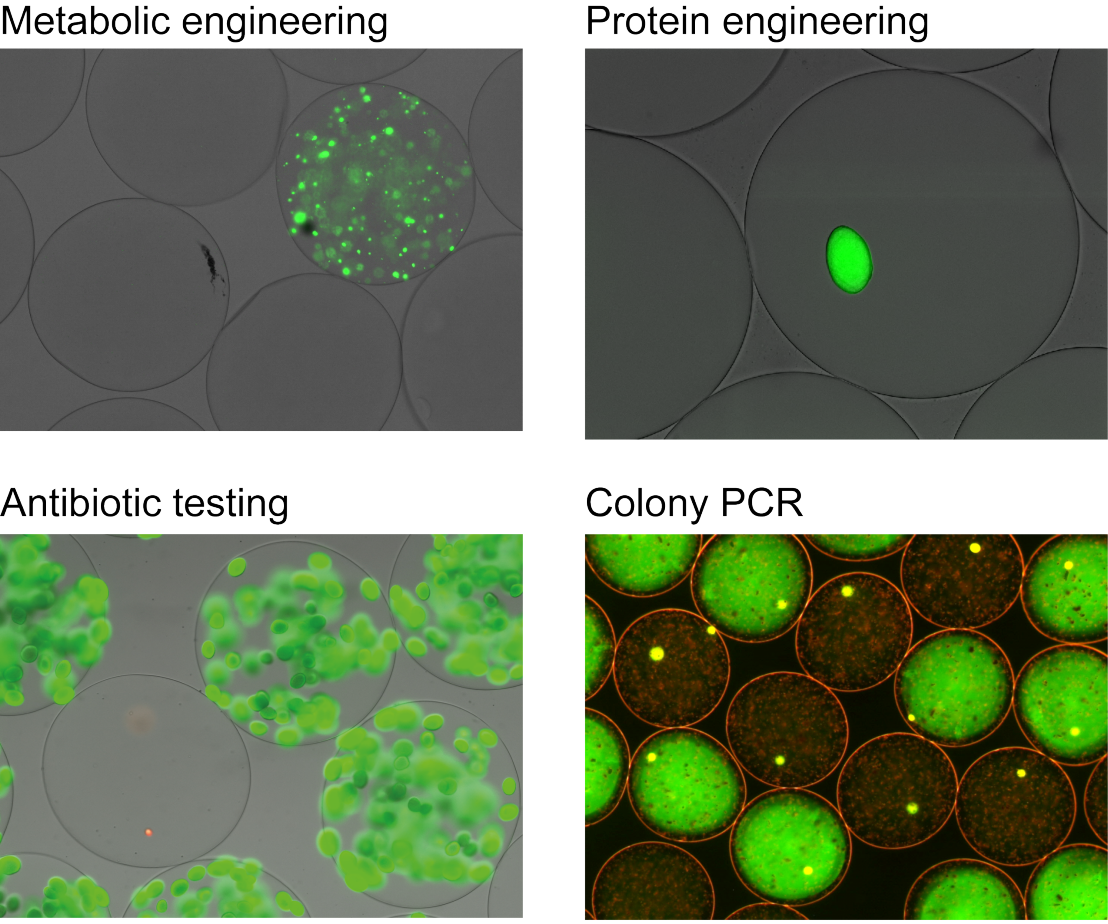
Nanoliter reactors (nLR) are monodisperse, nL-sized gel-particles used as reaction vessels for growth of mammalian or bacterial cells to microcolonies followed by analysis via flow-cytometry at a throughput of up to 1 million events per day.
Recently, we developed nLR assays for screening for improved vitamin producers generated by metabolic engineering, antimicrobial activity testing, isolation of improved proteins generated by directed evolution and specific DNA fragments.
References
Walser, M., Leibundgut, R. M., Pellaux, R., Panke, S. & Held, M.: Isolation of monoclonal microcarriers colonized by fluorescent E. coli, Cytom. Part A 73A, 2008, external page DOI.
Walser, M. et al.: Novel method for high-throughput colony PCR screening in nanoliter-reactors, Nucleic Acids Res. 37, 2009, external page DOI.
Meyer, A. et al.: Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors, Nat. Chem. 7, 2015, external page DOI.
Roberts, T. M. et al.: Identification and characterisation of a pH-stable GFP, Sci. Rep. 6, 2016, external page DOI.
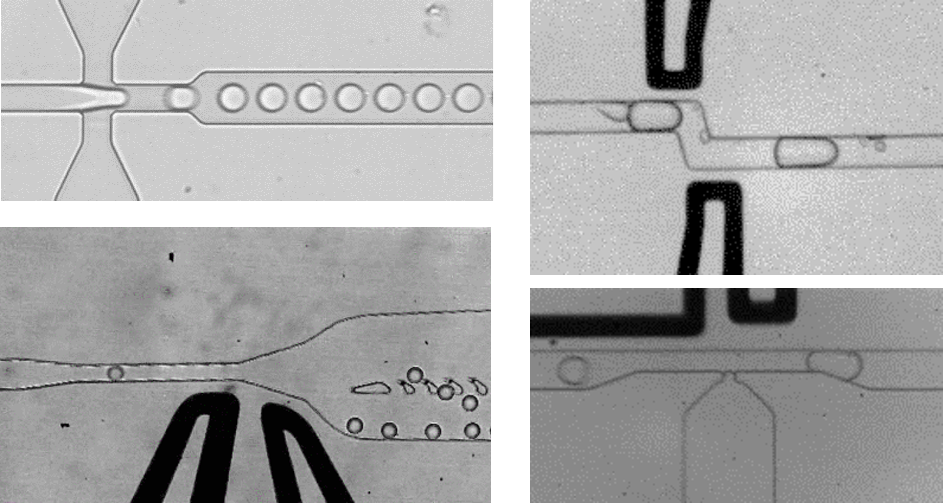
With the advent of droplet-based microfluidics, millions of compartments in the picoliter scale (spheres of < 50 µm diameter) can be produced in less than one hour. Advanced microfluidics allows manipulating each individual compartment, which gives us the opportunity to tweak the reaction conditions in multiple ways and conduct millions of different experiments at the same time. Compartments are subjected to injections, fusions, incubations and, in the final step, sorting in response to sensing of desired properties. These picoliter reactors can be analyzed and screened for example for improved protein properties in the range of 107 per day.
References
P. Rottmann, T. Ward, S. Panke: Compartmentalization – A Prerequisite for Maintaining and Changing an Identity, Chimia 70, 2016, external page DOI.

We employ single microbial cells (dimensions of 2 µm, total volume of roughly 1 to 2 fL) as hosts for transferring a genotype into a phenotype. Interesting properties are coupled to fluorescent signals that can be read out via flow cytometry, enabling a throughput of up to 109 “samples” per day.
We use these single-cell analysis assays to develop and improve biosensors or enzymes for increased catalytic activity.
Highly parallelized separation
The frequency at which libraries are sampled is still the rate-limiting step in high-throughput experimentation. We have developed a method for space- and time-efficient processing of very large sample numbers (107 per batch) and using nanoliter reactors (nLRs) as carriers. The protocol relies on gas-formation of nLR embedded microbes as a marker and buoyancy as readout. We use the method for both screening and selection protocols of recombinant microbes, generated by metabolic engineering. The system allows for phenotypic characterization and isolation of putative hits in a single operation, enabling large sampling rates exceeding 105 Hz (or >109 samples per day).

