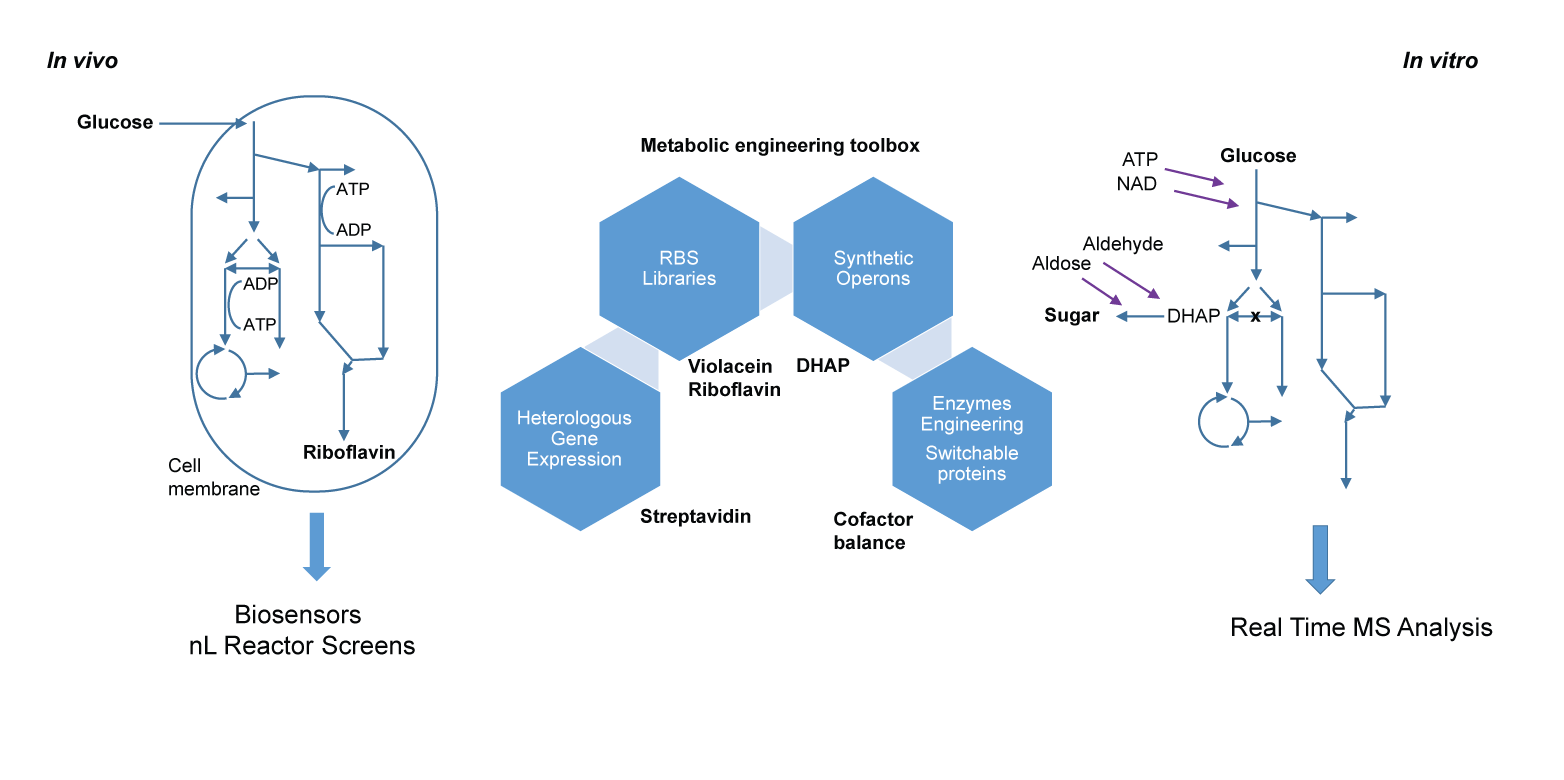
Multi-reaction systems
Metabolic engineering of biological systems aims to optimize conversion of carbon sources such as glucose to value added compounds. Our group makes use of living cells (in vivo) and cell-free systems (in vitro). In those systems we optimize the metabolite flux towards the product of interest, for example by knocking-out or down regulating enzymes responsible for unwanted side reactions. We do this by overexpressing enzymes of interest for product formation, by introducing heterologous genes that allow the catalysis of novel chemical reactions, or by engineering of enzymes in the pathway.

In vivo (whole cell) metabolic engineering has the advantage that cofactors are continuously regenerated and complex pathways are already present. Microbial systems often require the fine tuning of expression levels of endogenous or heterologous enzymes. We recently developed a ribosome binding site (RBS) engineering toolbox that has been utilized for the heterologous production of violacein and for streptavidin production in E. coli.
Reducing the enzyme level of essential proteins is non-trivial. As such, to avoid engineering at the genome level, we sometimes knock down their function at translational level by expressing small RNAs that sequester the ribosome binding site of the open reading frame to fine-tune the level to which gene expression is knocked down.
In addition to tweaking the enzyme levels we also we develop novel strategies to extend the cellular uptake capabilities of bacteria in order to make non-natural compounds available as substrates for metabolic engineering approaches.
References
Oesterle, S., Wuethrich, I., Panke, S.: Toward Genome-Based Metabolic Engineering in Bacteria, Adv. Appl. Microbiol., 2017, external page DOI.
Jeschek, M., Gerngross, D., and Panke, S.: Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort, Nature communications, 2016, external page DOI.
Jeschek, M., et al.: Biotin-independent strains of Escherichia coli for enhanced streptavidin production, Metabolic Engineering, 2017, external page DOI.
Oesterle, S. et al.: Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli, Sci. Rep., 2017, external page DOI.
Kuenzl, T., et al.: Overcoming the membrane barrier: Recruitment of γ-glutamyl transferase for intracellular release of metabolic cargo from peptide vectors, Metabolic Engineering, 2017, external page DOI.
In vitro (cell-free extract) metabolic engineering involves the removal of the cell wall and membrane provides access to the enzymatic machinery of the cell, precise adjustment of reactants and enzymes, and direct monitoring not only of products but also virtually all intermediates. Furthermore, such systems allow for the production of cytotoxic compounds or the complete draining of important metabolic intermediates.
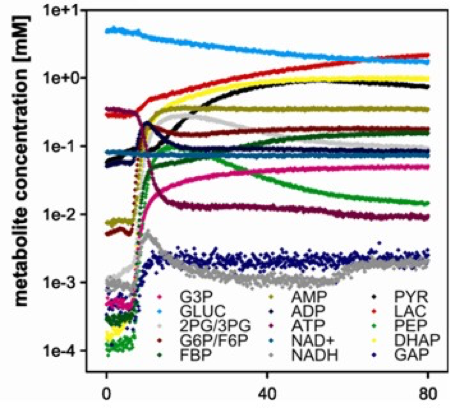
To monitor the dynamics of such processes and pinpoint the most promising points of intervention we developed mass spectrometry-based real-time analysis methods allowing for online measurement of the concentrations of multiple metabolites, for example in multi-enzyme systems employing an in vitro glycolysis to produce synthetic sugars.
Cell free extracts also contain a large number of enzymes whose activity can interfere with the cascade of interest. Some of them can be knocked out, however many cannot, e.g. if they were essential for making the cell-free extract in the first place. We make the corresponding enzymes “switchable”, which enables site-specific protease-catalyzed hydrolysis at the in vitro stage.
References
Bujara, M. et al.: Optimization of a blueprint for in vitro glycolysis by metabolic real-time analysis, Nat. Chem. Biol., 2011, external page DOI.
Oesterle, S. et al.: Sequence-based prediction of permissive stretches for internal protein tagging and knockdown, BMC Biology, 2017, external page DOI.