Xeno- and synthetic biology
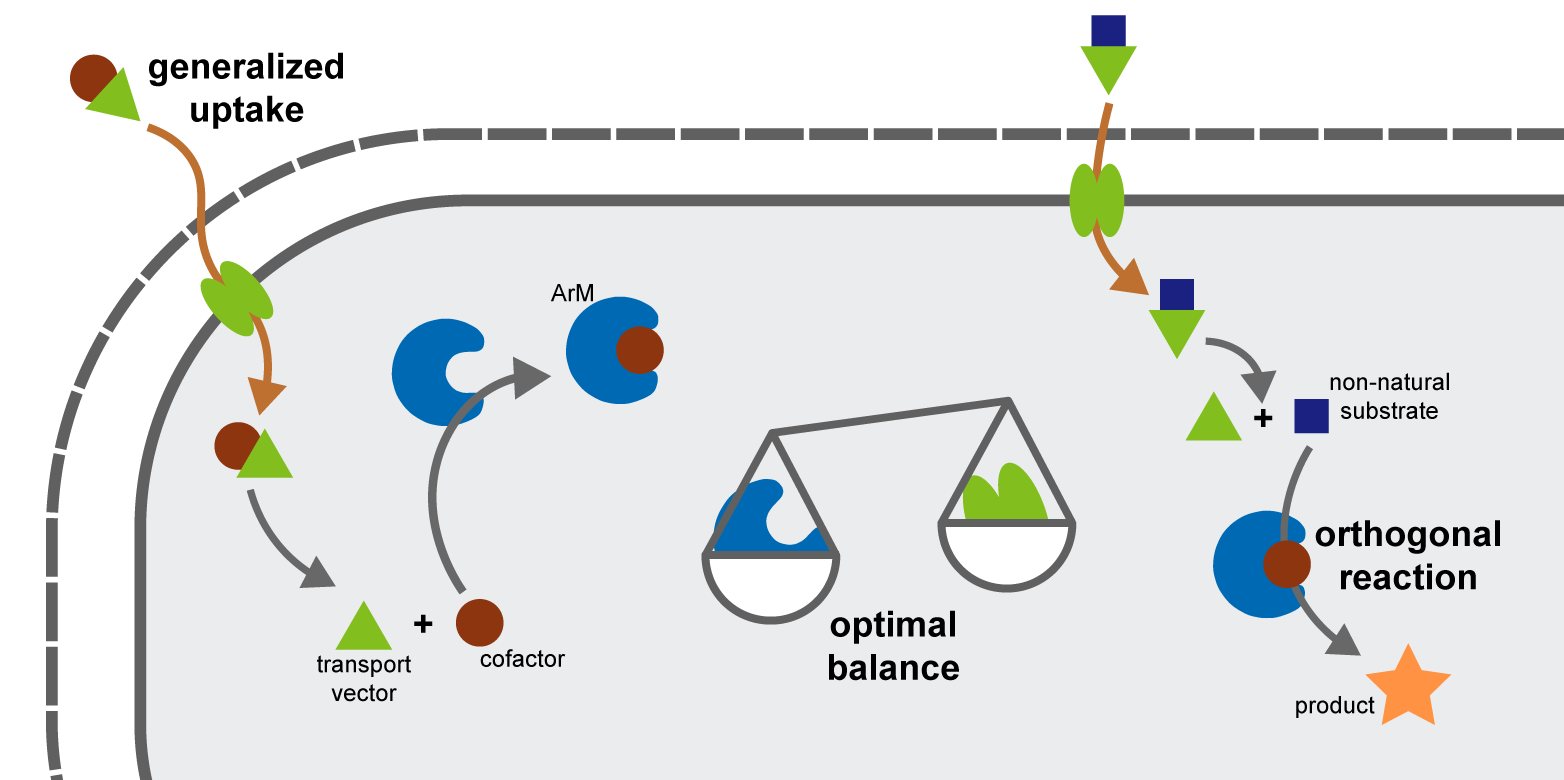
Our laboratory pursues different strategies to modify and extend features of organisms for the development of novel production hosts and intrinsically safe genetically engineered organisms, as well as to gain insights for basic research. Our main efforts revolve around the expansion of the biochemical repertoire of the cell by introducing bioorthogonal reaction mechanisms and the development of new strategies for the uptake of xenobiotic molecules that can be functionalized within the cell. Furthermore, we develop tools that allow smooth implementation of synthetic pathways into existing cellular metabolism and networks by finely balancing protein expression levels.
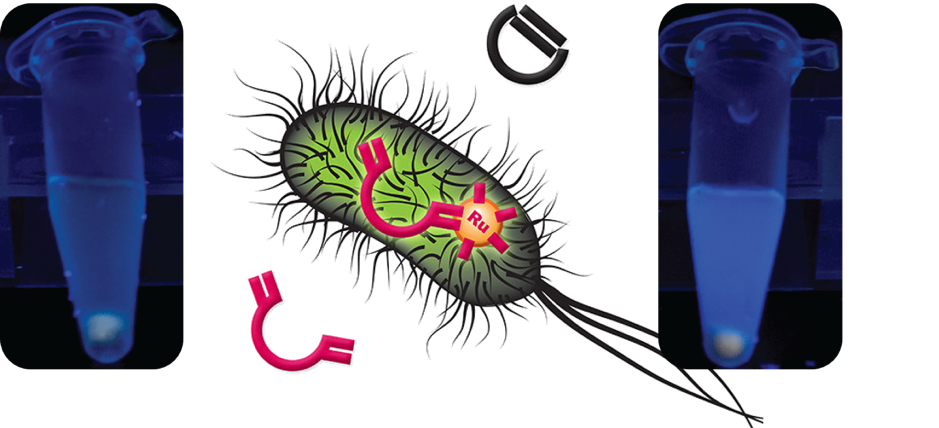
Our laboratory develops artificial metalloenzymes (ArMs) which contain artificial transition metal cofactors and therefore catalyze xenobiotic reactions not found in nature. Particular research interests in this context include the use of directed evolution to engineer ArMs in a high throughput fashion as well as the integration of these novel enzymes into the metabolic network of living cells. The latter constitutes a major bottleneck in the field of ArMs and represents an important step towards the establishment of new-to-nature metabolic pathways to produce hitherto inaccessible compounds in vivo using microbial cell factories.
References
Jeschek, M., Panke, S., & Ward, T. R.: Artificial metalloenzymes on the verge of new-to-nature metabolism, Trends in Biotechnology, 2017, external page DOI.
Jeschek, M., Reuter, R., Heinisch, T., Trindler, C., Klehr, J., Panke, S., & Ward, T. R.: Directed evolution of artificial metalloenzymes for in vivo metathesis, Nature, 2016, external page DOI.
Jeschek, M., Panke, S., & Ward, T. R.: Periplasmic screening for artificial metalloenzymes, Methods in Enzymology, 2016, external page DOI.

A frequent limitation when implementing new strategies in the field of xeno- or synthetic biology is the insufficient cellular uptake of foreign molecules that are neither recognized by endogenous transporters, nor have the properties to passively diffuse across the membrane. To overcome these limitations and thereby enable new applications in synthetic biology and metabolic engineering, we develop generalized solutions to expand the uptake capabilities of bacteria. For that purpose, we use a Trojan horse strategy in which we exploit the promiscuity of existing transport proteins by attaching cargo molecules to compounds (transport vectors) that are readily accepted by the transporter and develop enzymes to release the cargo inside the cell to make it available for intracellular applications.
References
Kuenzl, T., Sroka, M., Srivastava, P., Herdewijn, P., Marlière, P., Panke, S.: Overcoming the membrane barrier: Recruitment of γ-glutamyl transferase for intracellular release of metabolic cargo from peptide vectors, Metabolic Engineering, 2017, external page DOI.

Combining single pathway steps and integrating them into a production host remains challenging. To address this bottleneck, our laboratory is working on methods to precisely control protein levels by designing smart libraries that cover the multidimensional expression level space with minimal experimental effort. This allows us to finely balance multi-component metabolic pathways to increase productivity and minimize negative effects such as toxicity or metabolic burden. Moreover, these tools can be used to optimize expression of multi-protein complexes.
References
Jeschek, M., Gerngross, D. and Panke, S.: Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort, Nature Communications, 2016, external page DOI.
Jeschek, M., Gerngross, D. and Panke, S.: Combinatorial pathway optimization for streamlined metabolic engineering, Current Opinion in Biotechnology, 2017, external page DOI.