Programmable Manipulation of Cells
A team led by ETH Zurich Professor Yaakov Benenson has developed a Synthetic Biology platform that senses intracellular activities of mammalian transcription factors. This platform opens the way to precise sensing of and responding to diverse cell states and activities.
For millennia humans have been altering the genetic code of plants and animals by selectively breeding individuals with desirable features. As scientists have learned more about reading and manipulating the genetic code, they started transferring genetic information associated with useful features from one organism to another. Recent advances in Synthetic Biology have enabled bio-engineers to design multiple new DNA sequences from scratch. By combining these advances with engineering principles, synthetic biologists are now able to design cells and even organisms with new features.
Biology meets engineering
The interdisciplinary nature of Synthetic Biology makes it a particularly promising discipline, but the application of engineering principles to biological components creates challenges as well. As the engineering perspective is applied at all levels of biological structures - from molecules to cells, tissues and organisms - the redesign and construction of novel artificial biological pathways call for precise understanding of cellular processes. Consequently, one of the goals of Synthetic Biology is to develop programmable artificial gene networks that respond to endogenous molecular cues in order to analyse and understand cell behaviour. Transcription factors are an important class of such molecular cues due to their key role in determining cell identity and function. Yaakov (Kobi) Benenson, ETH Professor of Synthetic Biology at the Department of Biosystems Science and Engineering in Basel, and co-workers have developed a Synthetic Biology platform that is able to analyse and respond to cellular processes using endogenous transcriptional inputs. This programmable platform enables sensing and integrating multiple transcription factors, therefore leading to precise understanding of and response to diverse cell states and behaviors.
Cell machinery
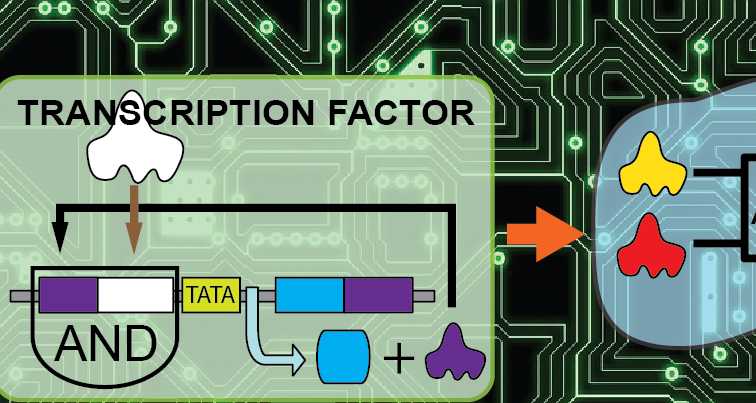
Every cell of a living organism contains an instruction set that determines its identity and function. These instructions are encoded in DNA: Complex molecular "strings" containing the genetic code, the so-called genome. The first step of decoding the genome, namely the transcription of genetic information from DNA to messenger RNA, is controlled by proteins called transcription factors. Numerous types of transcription factors interact to create the complex language of gene expression. Transcription factors perform this function by promoting or blocking the recruitment of RNA polymerase - an enzyme that transcribes genetic information from DNA to RNA - to specific genes. Bartolomeo Angelici, the project leader in the Benenson group, explains: "Transcription factors are proteins that have the ability to bind specific DNA sequences, switching the nearby genes ON or OFF, and determining which genomic instructions are carried out. Active genes are transcribed into mRNA and eventually translated into proteins, while inactive ones lay dormant." These so-called "expressed" genes determine cellular identity and behavior. He affirms: "This is why sensing combinations of active transcription factors is a powerful way to recognise specific cell types."
Sensing transcriptional activity
Recently, Angelici, Benenson and co-workers have established a framework for systematic design of selective and robust sensing, integration and transduction of transcriptional activity in mammalian cells. "The idea is to build synthetic gene circuits that sense specific transcriptional activities and are further wired to various downstream components. In this way, endogenous transcription factors can be rewired to control diverse processes in a programmable fashion", adds Benenson. "In order to sense transcriptional activity, we take advantage of the transcription factors’ ability to bind specific DNA sequences." Each minimal sensor is a DNA molecule containing the response element recognised by a given transcription factor, followed by one or more genes that are further wired to additional engineered components. Importantly, one of these genes is an artificial transcription factor that also serves as a sensor input, in what is known as "positive feedback" mechanism. Thus, after initial sensor induction by an endogenous factor, the artificial regulator is produced, amplifying its own amount and the amount of additional gene products.
Determining cell identity
Precise control of gene expression is a long-standing goal of Biotechnology and Biomedicine. Benenson’s new Synthetic Biology platform not only allows processing signals from multiple transcription factors and sensing molecular cues, but also responding with biologically active outcomes in a controlled fashion. For example, the platform might enable precise targeting of cells with a highly specific transcriptional profile. This is particularly interesting in complex diseases like cancer, because cancer cells are known to harbor abnormal transcriptional activities that distinguish them from healthy cells. In the future, a synthetic logic circuit sensing transcriptional factors could be used to specifically target cancer cells without harming healthy tissues.
Reference
Angelici B, Mailand E, Haefliger B, Benenson Y. Synthetic Biology Platform for Sensing and Integrating Endogenous Transcriptional Inputs in Mammalian Cells. Cell Reports 2016. doi: external page http://dx.doi.org/10.1016/j.celrep.2016.07.061