Computational tool: From single cells to tissues and whole organisms
Organisms and tissues develop through rounds of cell division, differentiation and death. Sophie Seidel and Tanja Stadler from the Computational Evolution group devised a new computational tool that quantifies these developmental processes from genetic footprints in single cells. Their work may contribute to a deeper understanding of development of model organisms and tissues, which in turn, can advance our insights into human development.
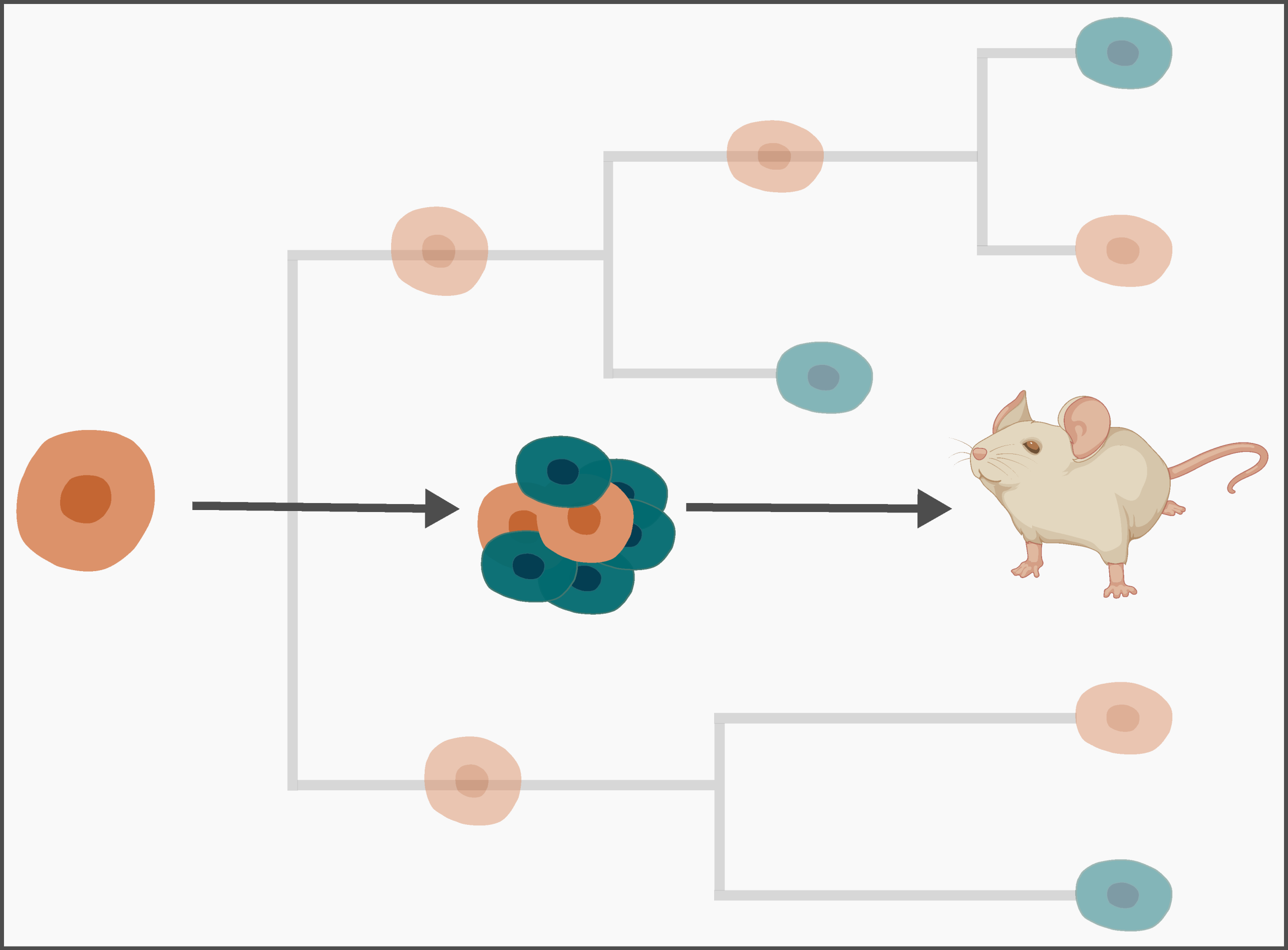
Find original paper:
Seidel, S and T Stadler (2022) TiDeTree: A Bayesian phylogenetic framework to estimate single-cell trees and population dynamic parameters from genetic lineage tracing data. Proceedings of the Royal Society B. external page https://doi.org/10.1098/rspb.2022.1844
Learn about the Computational Evolution lab led by Tanja Stadler.