A leap forward for computer simulations of cellular tissues
Many aspects of how cells organise in a tissue are still a mystery. Researchers from the Computational Biology group of Dagmar Iber use computer modelling to unravel the cellular organisation that gives rise to tissue shape and function in development and disease. A new powerful computer programme presented in Nature Computational Science is a big leap forward. The team hopes to be able to dig much deeper into the biophysics of the fundamental building blocks of life than was possible before.
The Computational Biology group of Dagmar Iber studies how cells work together to shape tissues during embryonic development, and how their misbehaviour can lead to diseases like carcinomas. With modern microscopy techniques, much can be learned about the size and shape of epithelial cells and their role in tissue morphogenesis. To fully understand the physical principles driving morphogenetic processes, biological hypotheses must, however, be formulated mathematically and then tested in computer simulations with data — a discipline the group specialises in.
Using such computer models not only allows the researchers to test hypotheses derived from microscopy imaging, but also to explore what is biologically or physically possible and what isn’t, by changing model parameters such as the biomechanical properties of the cells: how soft they are, how strongly they adhere to one another, or how fast they grow. Modifying such behaviour is usually very difficult or even impossible in an experiment on real tissue, but it’s often easy enough to do in computer simulations. Experimental and computational work thus go hand in hand, one motivating the other.
But biology is complex — more complex than most computer models are. Epithelial cells, that line all body surfaces, are polarised for instance: they have different sides with different properties, a feature that is essential for the many functions epithelia must fulfil in a viable organism. Their shapes are also vastly different, ranging from flat, squamous over cuboidal to highly elongated shapes, which makes it challenging to represent them in computer simulations. To top it off, there are typically thousands to millions of them in biological tissues. Existing computer programs that seek to simulate the mechanics of cellular systems are typically limited to either overly simplistic cell shapes such as spheres, or to small cell numbers. Even with modern computer power, the dynamics of epithelial tissues with realistic cell shapes and numbers has been too expensive to simulate. That’s where Dagmar Iber and her team saw the need for better, more powerful software.
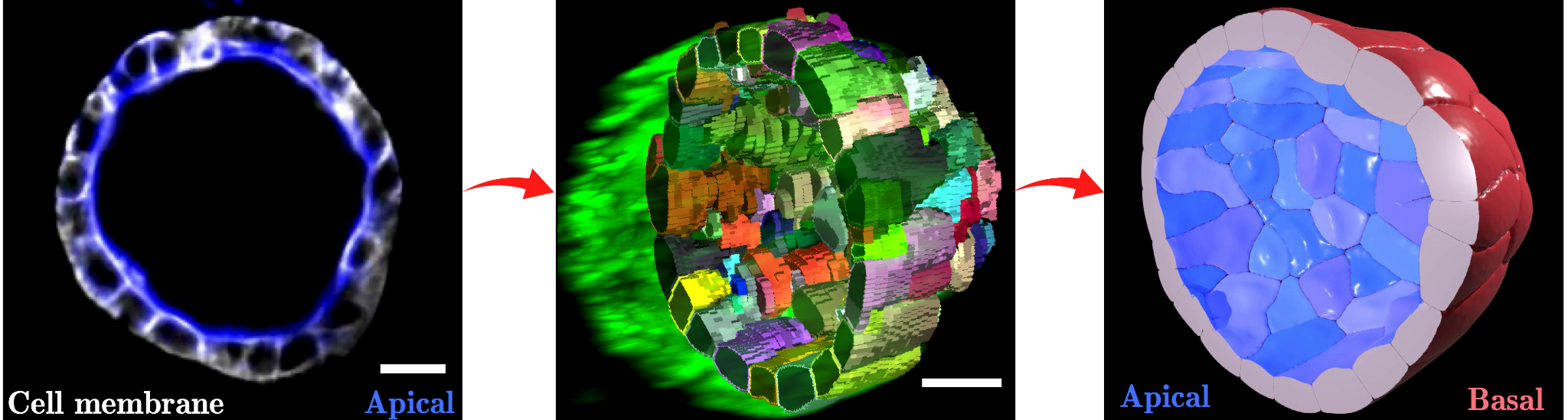
Four years ago, Steve Runser, a doctoral student in the research group, decided to take on the challenge of taking these computer simulations to the next level, bringing together what’s needed to address open questions in tissue development and disease: A highly detailed geometrical representation of cells, and the computational speed to simulate many thousands of them at the same time. The result of dedication and hard work: SimuCell3D, a new open source programme that has now appeared in Nature Computational Science, a renowned journal that publishes groundbreaking new computational tools and methods across all fields of science.
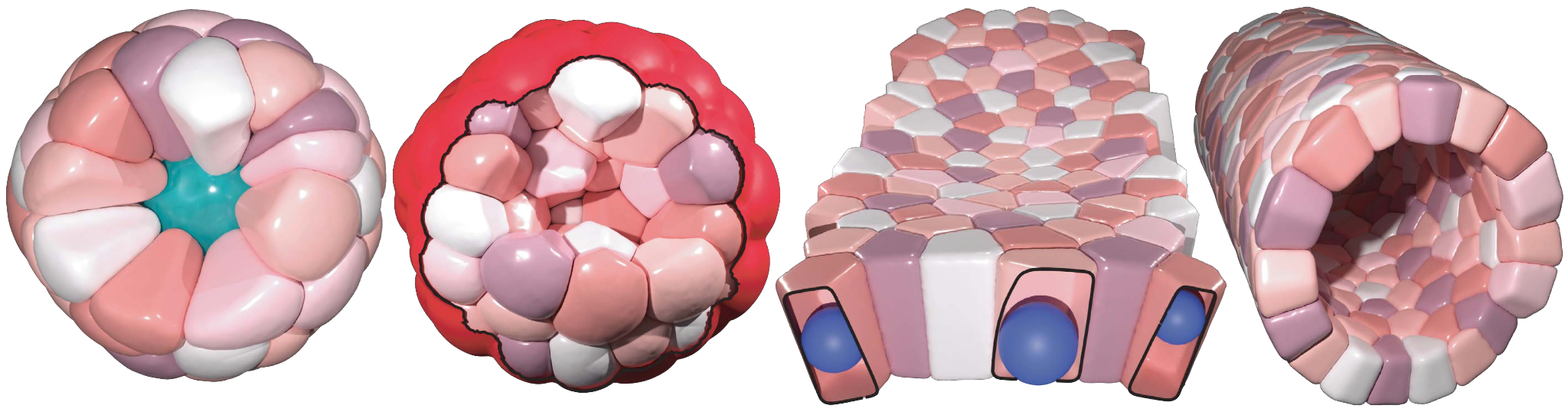
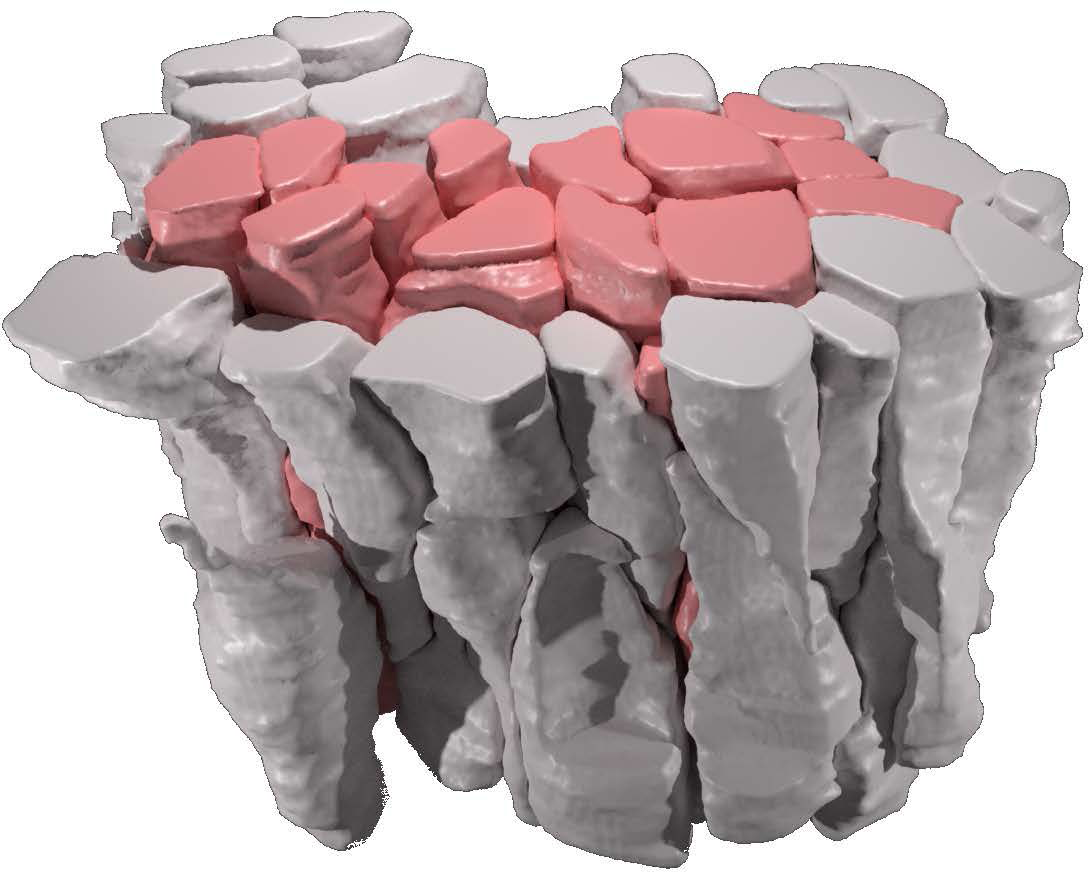
What’s special about SimuCell3D is that it combines two ingredients that are important for future simulations of the biomechanics of epithelia in one tool: First, it is much more efficient than previous programmes, allowing for the simulation of the dynamic deformation of dozens of thousands of cells with arbitrary shapes. This makes it possible for the first time to study large tissues in 3D with realistic cellular resolution. Secondly, the degree of biological detail with which the cells and their environment can be represented in SimuCell3D is unprecedented: No other comparable software previously allowed to include the nuclei of cells, the extracellular matrix, and cell polarity at the same time. Such features are important to account for, as they are expected to play a vital role in various morphogenetic processes.

SimuCell3D now becomes available to everyone around the globe for free as publicly available open source code. The developers hope that this will spur on a wide range of applications, spanning research on cell and developmental biology, immunology, cancer biology and beyond. The researchers in the Computational Biology group at D-BSSE look forward to seeing its many uses beyond their own research in tissue and organ development.
Find original publication in Nature Computational Science:
Runser, S, R Vetter, and D Iber (2024) external page SimuCell3D: three-dimensional simulation of tissue mechanics with cell polarization. Nature Computational Science, https://doi.org/10.1038/s43588-024-00620-9
Find the external page software for download: DOI: 10.5281/zenodo.10796908
Learn about the Computational Biology group of Dagmar Iber.