Antimicrobial resistance may evolve more rapidly than previously thought
As antimicrobial resistance increases globally, infections with resistant bacteria pose a major public health threat. In a new study in Nature Catalysis, a team of researchers led by Adrian Bunzel from the Bioprocess Laboratory at D-BSSE and Christopher Frohlich from the Arctic University of Norway revealed a novel mechanism for how resistances can emerge alarmingly fast. The discovery may help create antibiotics that are less prone to resistance and guide personalized health care.
Due to the widespread overuse of antibiotics in agriculture and healthcare, bacteria are often exposed to these drugs, allowing them to develop resistance. Resistance can develop by evolution through random mutations and natural selection. The ability to predict how bacteria evolve would enable the design of drugs that evade resistance and improve treatment strategies for patients. Unfortunately, predicting how mutations enhance resistance is extremely challenging. This is partly because mutations often behave unforeseeable – their effect on resistance development can differ when combined with other mutations. This phenomenon is described as epistasis.
To understand how epistasis can give rise to new mutations, the researchers replayed the evolution of a resistance-mediating enzyme in the laboratory. To that end, they exposed bacteria to a β-lactam antibiotic and monitored how their β-lactamase enzymes evolved to break down the drug. By studying the mutations that were introduced during evolution, they now reveal a thus far underappreciated mechanism of epistasis.
β-Lactam breakdown proceeds in several steps, involving binding of the drug to the β-lactamase and its subsequent cleavage. When studying the mutations introduced by evolution, Christopher Frohlich from UiT, the Arctic University of Norway, and Adrian Bunzel from the Bioprocess laboratory at D-BSSE discovered that the mutations accelerated either β-lactam binding or cleavage. Furthermore, their analysis of the native β-lactamase revealed that binding was slower than cleavage. Thus, mutations accelerating cleavage had little effect on the ancestral enzyme. In contrast, once a mutation was introduced that accelerated binding, mutations accelerating cleavage could greatly enhance resistance. This dependence of mutations on each other caused high epistasis, and shifting the slow step from binding to breakdown led to massive and unexpected increases in β-lactam resistance.
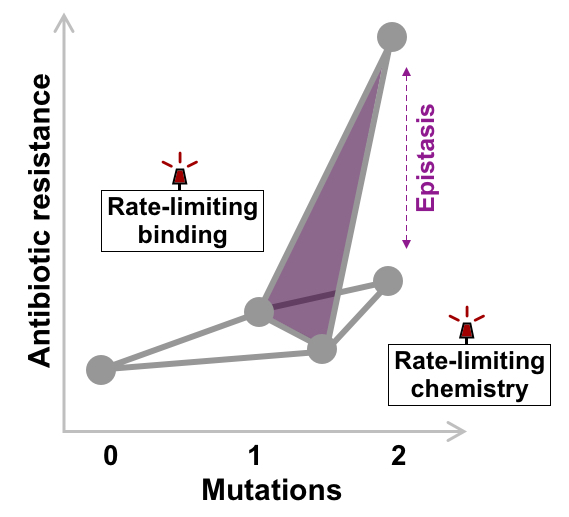
“If antibiotic binding is really slow, improving the chemical reaction is useless. But combining a mutation that improves chemistry with a mutation that accelerates binding can provide unexpectedly high improvements in activity,” says Adrian Bunzel.
Christopher Frohlich adds: “Understanding how resistance evolves is an important step in combating antimicrobial resistance. Our discovery will help to design more efficient antibiotics and could help guide personalized health care to treat infectious disease.”
Find the research article in Nature Catalysis:
C Frohlich, HA Bunzel, AJ Mulholland, MW van der Kamp, PJ Johnsen, HKS Leiros, and N Tokuriki (2024) Epistasis arises from shifting the rate-limiting step during enzyme evolution of a β-lactamase. Nature Catalysis, external page https://doi.org/10.1038/s41929-024-01117-4
Learn more about the research groups of Adrian Bunzel, D-BSSE/ETH Zurich, and external page Christopher Frohlich, UiT.