Novel computational approach to identify intracellular reaction processes
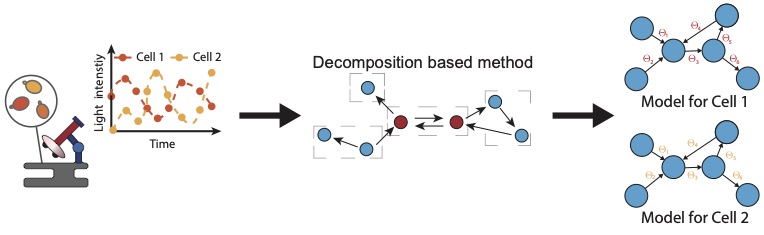
Intracellular processes, steered by the gene expression system, are high-dimensional and often intrinsically noisy. Advanced computational analysis of single-cell time-lapse microscopy data has the potential to unravel complex molecular networks within cells. In Nature Communications, researchers from the group of Mustafa Khammash present a novel way to decompose a large-scale network identification problem into several smaller, manageable subproblems.
The novel approach is scalable and efficient, and it proved to outperform existing methods. Led by Zhou Fang, this strategy was successfully applied to infer the transcription system in yeast from experimental data.

“The work pioneered by Zhou and Gupta introduces several computational innovations that uncover detailed statistical information from single-cell data, offering a unique glimpse into the complex dynamics within cells.”Mustafa Khammash, D-BSSE, Control Theory and Systems Biology lab
Find original article published in Nature Communications:
Fang, Z, Gupta, A, Kumar, S, and M Khammash (2024) Advanced methods for gene network identification and noise decomposition from single-cell data. external page Nature Communications, external page https://doi.org/10.1038/s41467-024-49177-1
Learn about research in the Control Theory and Systems Biology lab led by Mustafa Khammash.