Unveiling the mechanisms underlying human cell diversity
How do cells with the same genetic material differentiate into diverse cell types during early embryonic development? In a study in Nature Neuroscience led by the Quantitative Developmental Biology group of Barbara Treutlein in collaboration with colleagues from the Roche Institute of Human Biology, scientists explore at single-cell level the role of histones in serving as on-/off-switches for gene expression during cell development and differentiation in brain organoids.
During development and differentiation, cells undergo a remarkable transition from the pluripotent stem cell to the differentiated cell that fulfils diverse functions in the human body. Histone proteins – and modifications to their structures – play a key role during this differentiation process as epigenetic markers, determining which genes on the DNA are activated for read-out or deactivated and their expression repressed. Despite recent progress in the field of epigenetics, the exact mechanisms behind gene expression in brain development remained elusive.
Researchers from the Quantitative Developmental Biology group of Barbara Treutlein and colleagues from the group of Gray Camp at the Roche Institute of Human Biology now shed light on how human stem cells acquire and maintain their identity as neuronal and glia cells. They used brain and retina organoids – miniature, simplified versions of organs grown from stem cells in the lab – as model systems that mimic the formation and differentiation of neural tissue.
Researchers from the Quantitative Developmental Biology group of Barbara Treutlein and colleagues from the group of Gray Camp at the Roche Institute of Human Biology now shed light on how human stem cells acquire and maintain their identity as neuronal and glia cells. They used brain and retina organoids – miniature, simplified versions of organs grown from stem cells in the lab – as model systems that mimic the formation and differentiation of neural tissue.
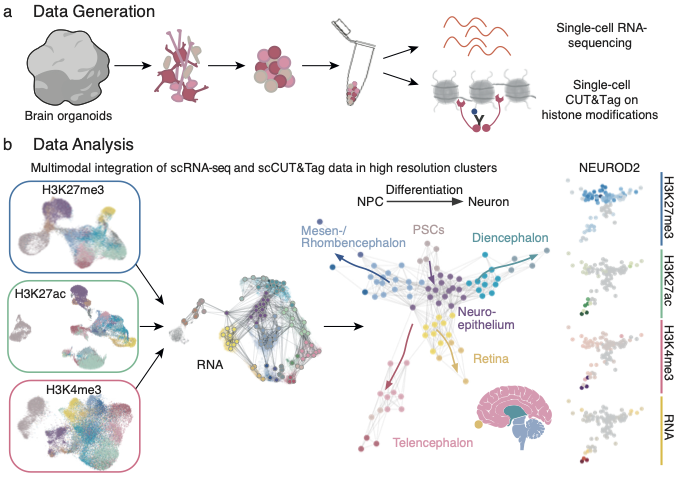
In a study published by Nature Neuroscience, the team focused on three different modifications of the histone protein H3. One modification was linked to gene repression, one modification associated with the active expression of genes and one modification found at active enhancers for gene expression. The researchers utilized a technique called single-cell CUT&Tag to pinpoint these histone modifications in individual cells and combined this data with gene expression profiles. Advanced computational methods were then used to integrate the epigenetic and transcriptomic data, creating an atlas that traces the progression of cells from pluripotency to specialized brain cells.

“This atlas allows us to follow at single-cell level the DNA and gene expression dynamics of each gene throughout stem cell differentiation into neural cell types of different brain organoid regions.”Barbara Treutlein, Head of the Quantitative Developmental Biology group at D-BSSE
One of the key findings of the study is that neuronal genes are primed for activation by specific histone modifications even before their expression increases, ensuring they are ready to be switched on when needed. Fides Zenk, first author of the study and since recently, group leader at the EPF Lausanne, emphasizes: “Our findings add to the growing evidence that histone modifications can have a functional role in directing gene expression and are not installed as a mere consequence of transcription”.
By mapping the epigenetic changes that occur during brain development, this study offers a detailed view of how pluripotent cells differentiate into specialised brain cells. The findings lay the foundation for future studies that aim at controlling histone modifications to direct cell fate and function, opening new avenues for regenerative medicine and the modelling of neurodevelopmental disorders.
Find the original paper in Nature Neuroscience
Zenk, F., Fleck, J.S., Jansen, S.M.J. et al. (2024) external page Single-cell epigenomic reconstruction of developmental trajectories from pluripotency in human neural organoid systems. Nature Neuroscience, https://doi.org/10.1038/s41593-024-01652-0
This paper is featured as external page Research Briefing.
Learn about the Quantitative Developmental Biology group led by Barbara Treutlein.